API Overview
The CGC API uses the REST architectural style to read and write information about projects on the CGC. The API can be used to integrate the CGC with other applications, and to automate most procedures on it, such as uploading files, querying metadata, and executing analyses.
The base path of the API is: https://cgc-api.sbgenomics.com/v2
On this page
API paths
The paths are structured into the following endpoints, which cover different categories of activity on the CGC:
General API information
Format
API requests are made over HTTP, and information is received and sent in JSON format. For this reason, you should set both the accept and the content header of the request to application/json.
Responses also include CGC-specific error codes, in addition to standard HTTP codes. Information about each code is available on the page API status codes.
Generic query parameters
All API calls take the optional query parameter fields. This parameter enables you to specify the fields you want to be returned when listing resources (e.g. all your projects) or getting details of a specific resource (e.g. a given project).
The fields parameter can be used in the following ways:
- No
fieldsparameter specified: calls return default fields. For calls that return complete details of a single resource, this is all their properties; for calls that list resources of a certain type, this is some default properties. - The
fieldsparameter can be set to a list of fields: for example, to return the fieldsid,nameandsizefor files in a project, you may issue the callGET /v2/files?project=john_doe/project1&fields=id,name,size. The same goes for a call to get details of a specific file. - The
fieldsparameter can be used to exclude a specific file: if you wish to omit certain field from the response, do so using thefieldsparameter with the prefix!: for example, to get the details of a file without listing its metadata, issue a callGET /v2/files/567890abc8a5136ec6127063?fields=!metadata. The entire metadata field will be removed from the response. - The
fieldsparameter can be used to include or omit certain nested fields, in the same way as listed in 2 and 3 above: for example, you can usemetadata.sample_idororigin.taskfor files. - To see all fields for a resource, specify
fields=_all. This returns all fields for each resource returned. Note that if you are getting the details of a specific resource, the use offields=_allwon't return any more properties than would have been shown without this parameter – the use case is instead for when you are listing details of many resources. Please use with care if your resource has particularly large fields; for example, therawfield for an app resource contains the complete CWL specification of the app which can result in bulky response if listing many apps. - Negations and nesting can be combined freely, so, for example, you can issue
GET /v2/files?fields=id,name,status,!metadata.library,!originorGET /v2/tasks?fields=!inputs,!outputs.
Identifying projects, users, apps, files, tasks and inputs
Project short names
Projects on the CGC have both given names, which you will see in visual interfaces, like the Projects drop-down menu on the visual interface, and short names, which are human-readable IDs derived from the given names. To refer to a project in an API call, you should use its short name.
Project short names are based on the name you give to a project when you create it. The short name is derived from the project name by:
- Formatting the name in lower case
- Omitting special characters, that are not letters, numbers, spaces or underscores
- Replacing spaces with hyphens
- Replacing underscores with hyphens
- Adding
_1to any name that is already assigned to one of your projects.
For example, if I give my project the name 'RFranklin's experiments', it would be automatically assigned the short name 'rfranklins-experiments'.
You can optionally override an auto-assigned short names to one of your choice, when you create a project. To create your own project short name, first create a project, using the drop-down menu at the top of the screen. Then, click the pencil icon on the Create a project pop-out window.
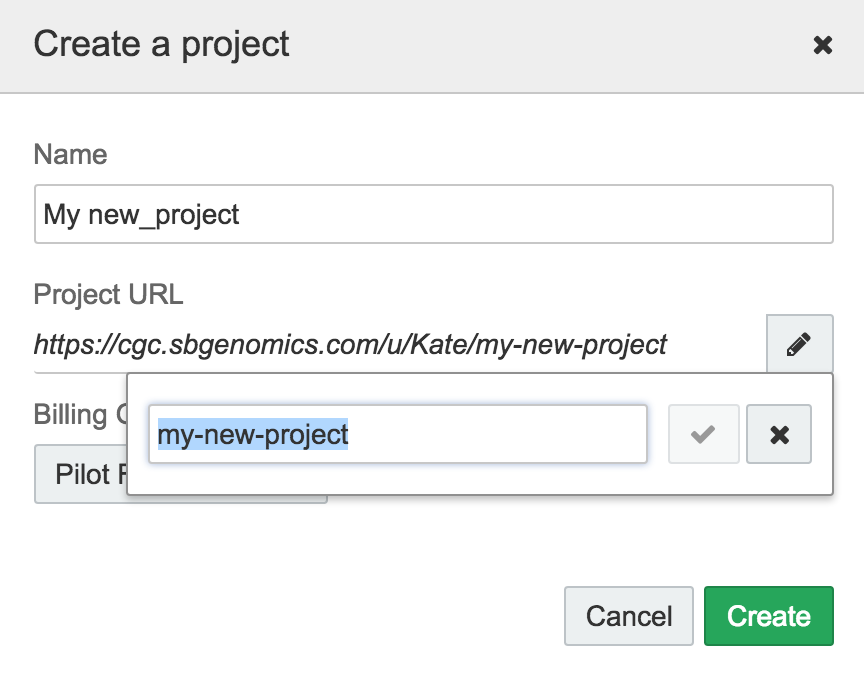
Click the pencil icon to edit the project short name.
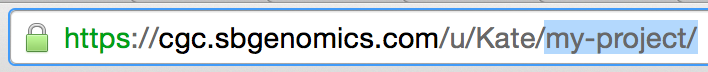
To check a project's short name, or a task or file's ID, you can inspect the URL when you click on the object in the browser.
Changing a project's name
Note that once the project has been created, you cannot change its short name. However, you can edit a project's given name at any time.
Users
CGC Users are referred to in the API by their usernames. These are chosen by the user at the point at which they sign up for the CGC. Usernames are unique and immutable. They are also case sensitive, so it is advisable to user lower case strings for your username to avoid ambiguity.
Uniqueness of project names
Every project is uniquely identified by
{project_owner_username}/{shortname}.
Apps
Apps (tools and workflows) in projects can be accessed using the API. Like projects, apps have both given names, which are assigned by the users who create them, and short names An app's short name is derived by the same process as a project's short name.
Each app is identified with reference to the project it is contained in and its short name, using the format: {project_owner}/{project}/{app_short_name}/{revision_number}.
For instance, RFranklin/my-project/bamtools-merge-2-4-0/0 identifies an app.
Tasks
Tasks are referred to in the API calls by IDs. These are hexadecimal strings (UUIDs) assigned to tasks. You can retrieve them by making the API call to list tasks.
Tasks have the following statuses: DRAFT, RUNNING, QUEUED, ABORTED, COMPLETED or FAILED.
Files
Files are referred to in API calls by IDs. These are hexadecimal strings assigned to files. You can retrieve them by making the API call to list files.
Note that file IDs are dependent on the project the file is stored in. If you copy a file to a different project, it will have a new ID in this project.
In calls that return CWL descriptions of tasks, such as the call to GET task details, files are identified by their path objects. The file path is identical to the file ID.
Inputs
Task inputs are specified as dictionaries. They pair apps to be executed in the task with the objects that will be inputted to them.
The format for an input is:
{app_id}: {object}
The {app_id} is defined above. The value of {object} is obtained as follows:
If the object to be inputted to the task is not a file (but an integer, boolean, etc) then simply enter that value as {object}.
If the object to be inputted to the task is a file, then {object} is a dictionary, with the format:
{
"class": "File",
"path": "file_id",
"name": "file_name.ext"
}
When multiple files are used as inputs, enter a list of {object}s, like this:
[
{
"class": "File",
"path": "file_id",
"name": "file_name.ext"
}
{
"class": "File",
"path": "file_id",
"name": "file_name.ext"
}
]
The following are all examples of inputs:
- An input integer:
"Offset": {2}
- An input file for the known indels:
{
"cuffdiff_zip": {
"class": "File",
"path": "567890abc9b0307bc0414164",
"name": "example_human_known_indels.vcf"
}
}
3: File inputs for a Whole Exome Sequencing workflow, in the form of FASTQ reads:
"Reads_FASTQ": [
{
"class": "File",
"path": "567890abc3d8130ea4047731",
"name": "WES_human_Illumina.pe_1.fastq"
},
{
"class": "File",
"path": "567890abc8a5136ec6127063",
"name": "WES_human_Illumina.pe_2.fastq"
}
]
Task inputs
For more examples of task
inputs, use the call to get task inputs for some of the tasks you initiate on the CGC visual interface.
For finding which app receives which inputs and their format, you can review the app's page on the CGC visual interface. For example Whole Exome Sequencing GATK 2.3.9.-lite
Authentication
To set your CGC credentials on the API, you will need an authentication token, which you can obtain from https://cgc.sbgenomics.com/account/#developer.
All API requests need to have the HTTP header X-SBG-Auth-Token which you should set to your authentication token. The only call which is exempt from this is the '/' call to list all request paths.
Rate limit
The API rate limit is a limit to the number of calls you can send to the Seven Bridges public API within a defined time frame (learn more).
Response pagination
All API calls take the pagination query parameters limit and offset to control the number of items returned in a response. These are useful if you are returning information about a resource with many items, such as a list of many files in a project.
Filtering
In addition to controlling the number of items returned using the pagination query parameters, if you are requesting information about files using the call to
GET /filesyou can filter items returned by filename, metadata, or originating task.
Specify the number of items to return in a response
You can control how many items are returned by an API call using the query parameter limit. If you do not specify a value for limit in a call, a maximum of 50 items will be returned by the call by default.
The maximum value for the query parameter limit is 100.
Example 1:
Suppose you have 70 files in the project my-project, and you issue the call to GET /files as follows:
GET /v2/files?project=my-project HTTP/1.1
Host: api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
Since no value for limit was specified, this call will return details of 50 of the files, along with a URL to return the next 20.
Example 2:
Again, suppose you have a project my-project with 70 files in it. The following call will return details of all 70 files"
GET /v2/files?project=my-project?limit=70 HTTP/1.1
Host: api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
Specify the starting point for items to return in a response
You can control the starting point at which to start returning items in an API call using the query parameter offset. If you do not specify a value for offset then the default starting point will be the first item in the specified resource. Specifying an integer value for offset will start from the item which is the one after the specified integer value.
Example 1:
Suppose you have a project called my-project containing 70 files, and you want to return their details, starting with the 31st file. To do this, issue the call to GET /files with a query parameter offset specified as follows:
GET /v2/files?project=my-project?offset=30 HTTP/1.1
Host: api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
Calls made with the offset query parameter additionally return the header X-Total-Matching-Query which signifies the total number of results.
Example 2:
An example of a call made using both pagination parameters is as follows:
GET v2/projects?limit=2&offset=2 HTTP/1.1
Host: api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
This returns the following body in JSON:
{
"href": "https://api.sbgenomics.com/v2/projects/",
"items": [
{
"href": "https://api.sbgenomics.com/v2/projects/john_doe/project1",
"id": "john_doe/project1",
"name": "project1"
},
{
"href": "https://api.sbgenomics.com/v2/projects/john_doe/project2",
"id": "john_doe/project2",
"name": "Project 2"
}
],
"links": [
{
"href": "http://api.sbgenomics.com/v2/projects/?offset=4?limit=2",
"rel": "next",
"method": "GET"
}
]
}
The headers returned include X-Total-Matching-Query which lists the total number of results.
The body of the response includes the array links, which indicate how to get the next or previous set of results.
Updated less than a minute ago
