Import data from the PDC
Latest available release of PDC data on the CGC corresponds to the PDC Data Release V2.6 of January 27, 2021. Get more information about updates of PDC data on the CGC.
About the PDC
The NCI Cancer Research Data Commons (CRDC) aims to create a scalable infrastructure that provides secure access to many different data types across scientific domains, allowing users to analyze, share, and store results, leveraging the storage and elastic compute of the cloud. As a node in this CRDC ecosystem, the Proteomic Data Commons (PDC) is a pilot project to democratize access to cancer-related proteomic datasets as well as to provide sustainable computational support to the cancer research community.1
The process of importing files from the PDC to the Cancer Genomics Cloud (CGC) consists of the following two stages:
- Downloading a manifest file from the PDC website.
- Importing files to the CGC based on the downloaded manifest file.
Downloading manifest files from the PDC
Some NCI data are under an EMBARGO for publication and/or citation until a specific date known as embargo date. Here's what characterizes such data on the CGC:
- Embargoed files will have an EMBARGOED label next to them.
- Embargo date is inherited. If you run an analysis using embargoed files as inputs, all output files resulting from the analysis will inherit the embargo date from the input files. If files with different embargo dates are used as inputs, all output files will inherit the embargo date that expires last.
- Embargo date is stored as a metadata field.
For more details, visit the NCI Proteomic Data Commons for the study of interest.
Manifest files that are downloaded from the PDC contain information about the data you want to import in the second stage of this process.
To download a manifest file from the PDC:
- Open the PDC website.
- Select the Files tab below the chart. A list of all files is displayed below.
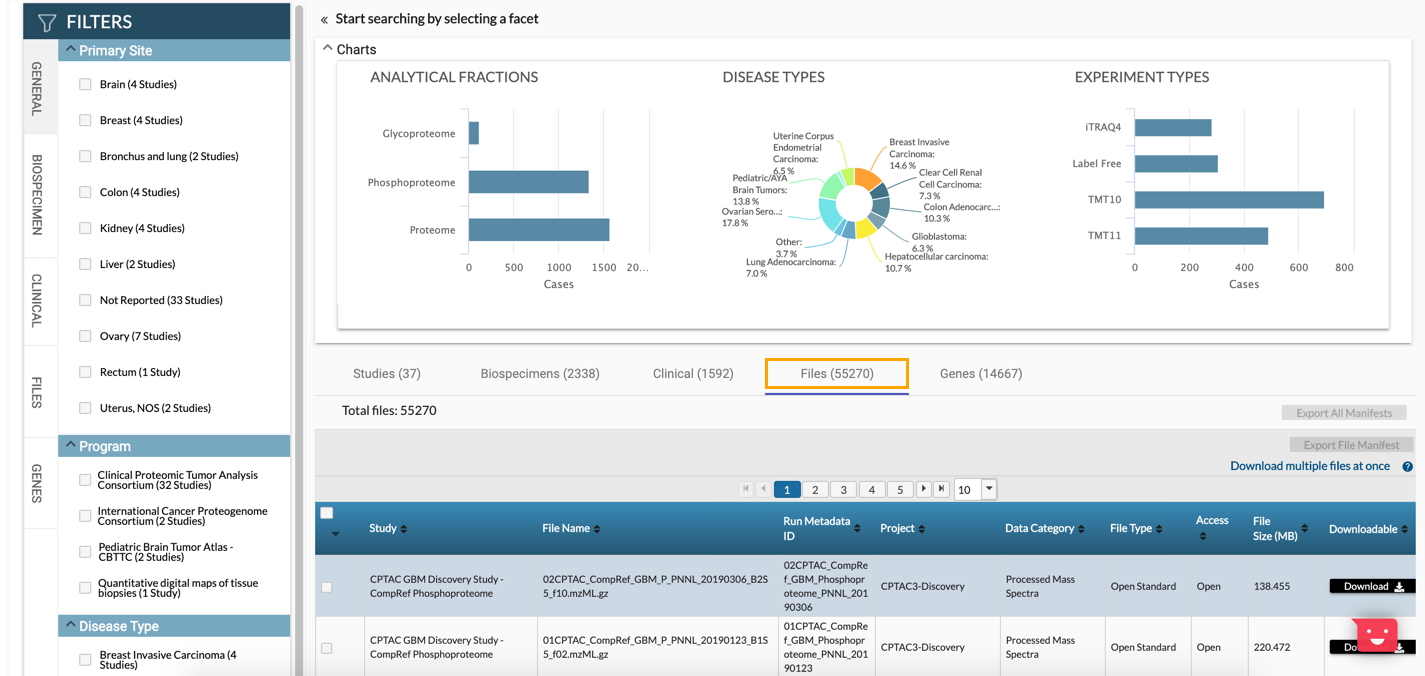
- (Optional) In the Filters pane, use the available filtering options to narrow down the search results.
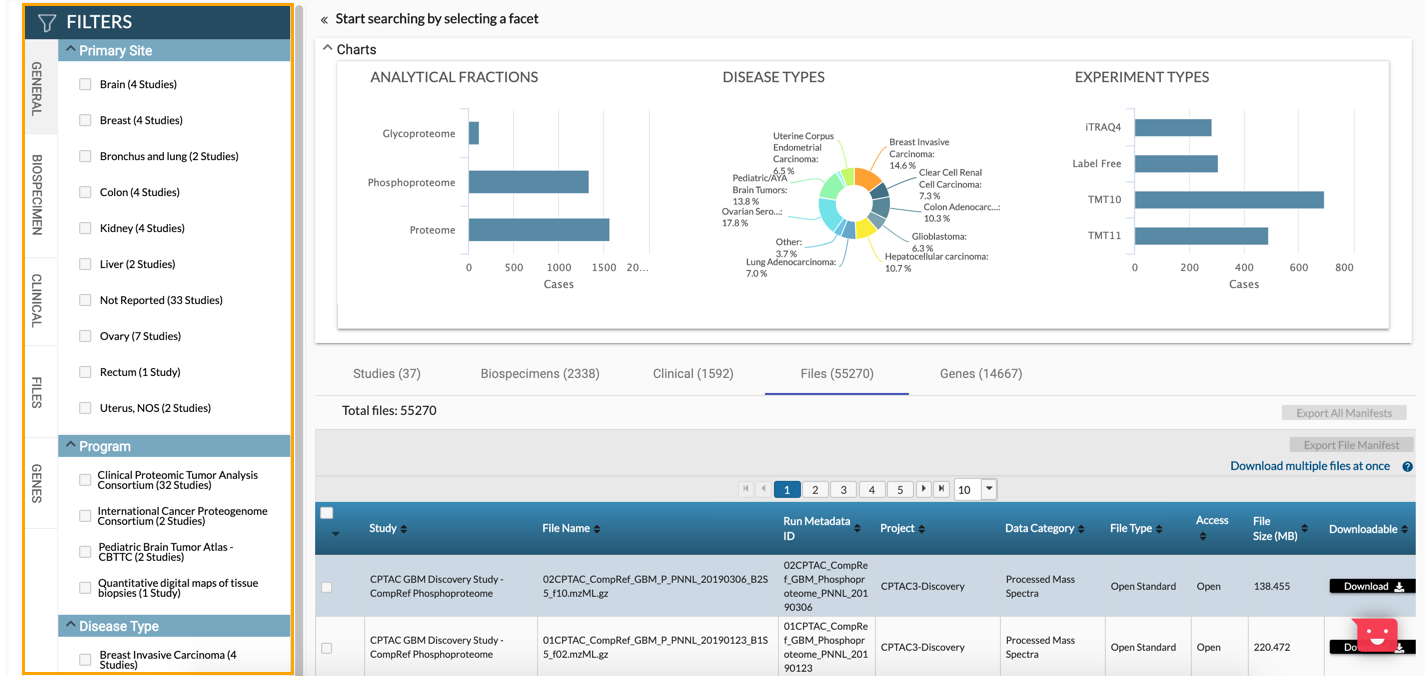
- Check the boxes next to the files you want to download.
- Click CSV next to Export File Manifest in the top-right corner above the table. A manifest file in the CSV format is downloaded to your computer. Please keep the file as it will be used in the following stage of the import process.
Import files from the PDC to the CGC
- Navigate to a project on the CGC.
- Once in the project, click the Files tab.
- Click Add files > Import from a manifest file.
- In the Import files from dropdown, select Proteomics Data Commons (PDC).
- Click Browse files and select the manifest file from your local machine, or drag and drop the file onto the marked area. Alternatively, if you have already uploaded your generated manifest file to a project, click Select manifest from project and select the file.
- (Optional) In the Add tags field, add the keywords (tags) that describe the imported items.
- Resolve naming conflicts - Select the action to be taken if a naming conflict occurs. Available actions are Skip and Auto Rename. Read more about naming conflicts resolution.
- Click Import. The file import process starts.
Updated less than a minute ago
