The Picard Alignment Summary Metrics application is a quality control tool for Exome-Seq and RNA-Seq alignments.
It compares an alignment file (either BAM or SAM) to the reference file (FASTA), and provides statistics to assess the alignment, such as the number of input reads and the percent of reads that are mapped.
An Alignment Summary Metrics report is available for all files with the extension .summary_metrics.txt.
Click any file with the extension .summary_metrics.txt to access its summary report.
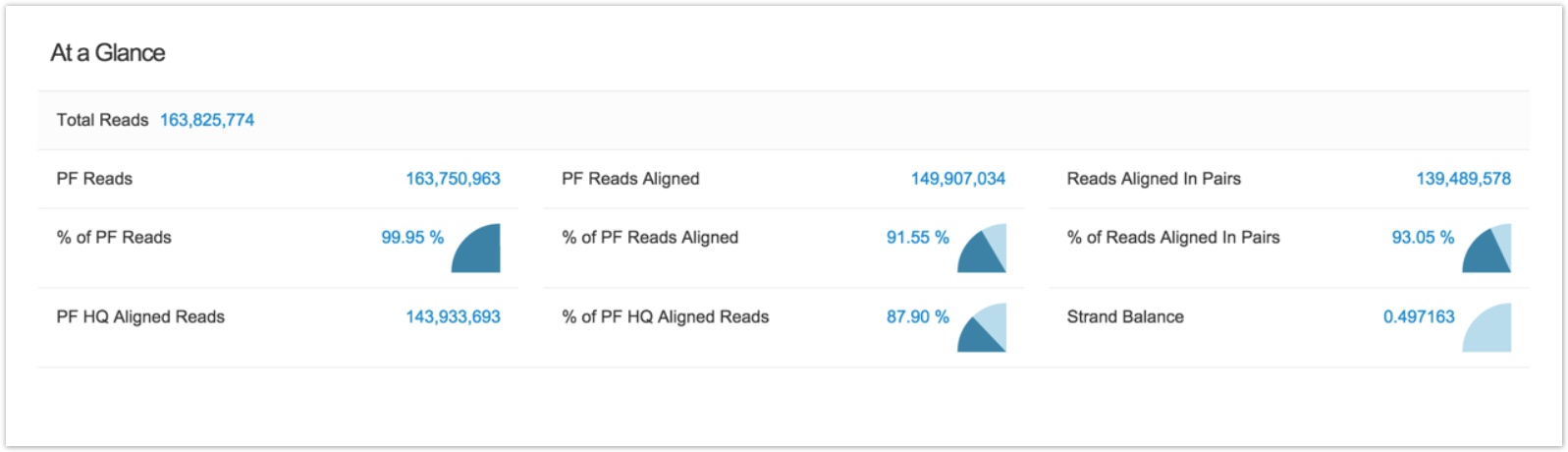
To see the full list of metrics reported by Alignment Summary Metrics , click Show details on its summary report.
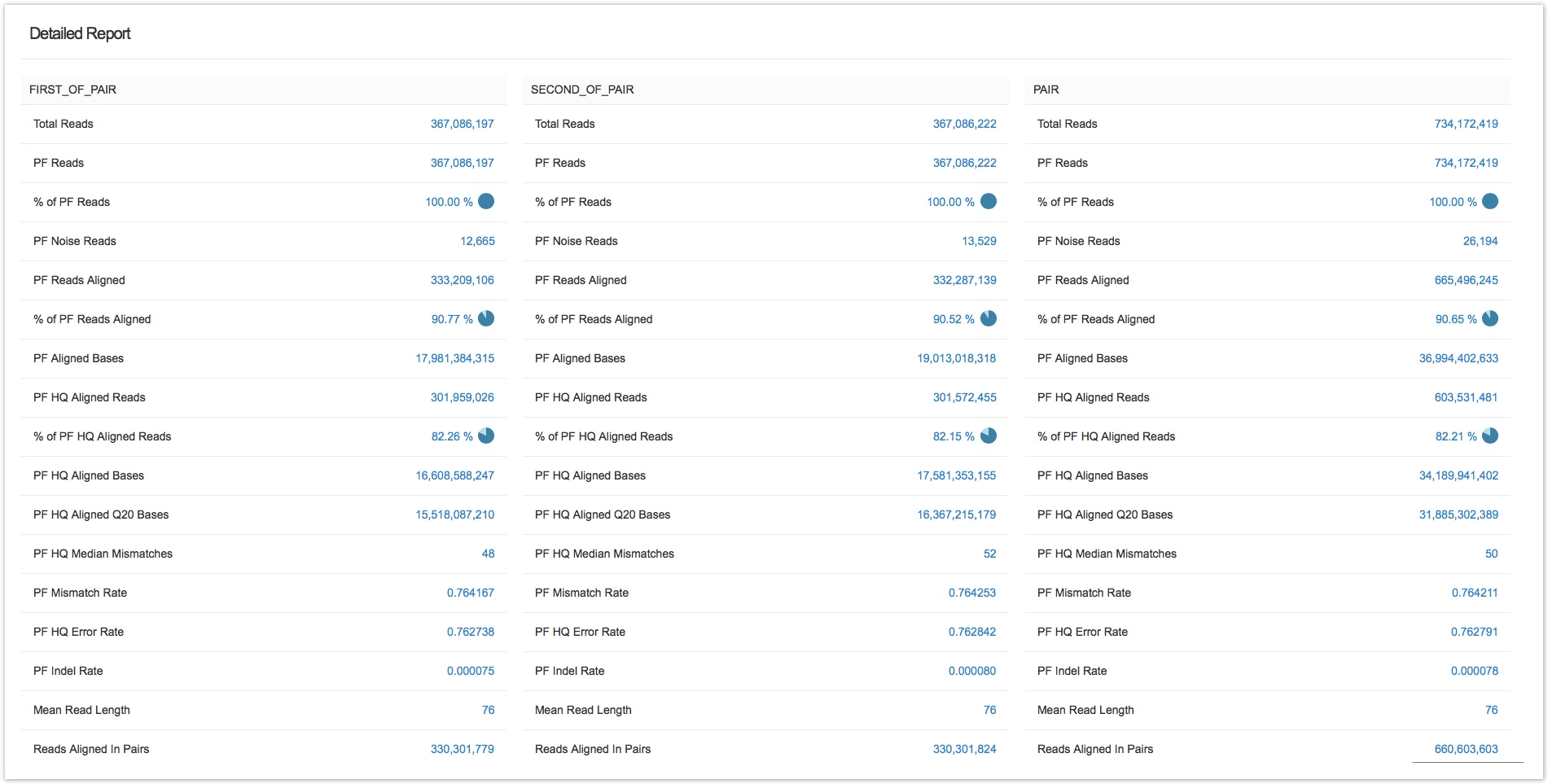
For a full explanation of the information shown in the report, we refer you to the package tutorial.
Updated over 3 years ago
