About batch analyses
Overview
Batching runs the same workflow or tool multiple times with varying inputs in parallel executions. These inputs can be grouped by input files or via specified metadata criteria.
Learn more about setting up a batch task. On this page, learn more about batching.
What are batch tasks?
Batch analysis separates files into batches or groups when running your analysis. The batching is done according to the specified metadata criteria of your input files or according to a files list you provide.
When your batch task is run, it produces multiple child tasks: one for each group of files. The child tasks perform the same analysis independently.
Running a batch task will produce a number of child tasks equal to the number of groups your inputs have been separated into. There are a series of advantages to running a batch task:
- The batch task automatically generates identical tasks to be run on your groups of files, saving you the effort of having to do this manually.
- Child tasks are executed in parallel and are independent from each other.
- If a child task fails, this does not affect the other child tasks, meaning that you only need to troubleshoot and rerun the failed task.
- You can re-run any child task separately.
What is metadata?
Metadata is information about the nature of your data and how it was obtained. This information is used by apps on the CGC to make sure the right data gets analyzed together. Learn more about supplying metadata for your files.
Batch tasks use values for metadata fields or file names in a given file list to group the files you are analyzing.
Some common scenarios include batching by:
- File name
- Case ID
- Sample ID
- Library ID
- Platform ID
- File segment
How do I run a batch task?
You can set up a batch analysis on the CGC both using the visual interface, as well as via the API.
Example of batching by metadata
Let's imagine having to run the RNA-seq Alignment - STAR workflow on many files.
The metadata fields by which batching is possible via the visual interface are described in the figure below.
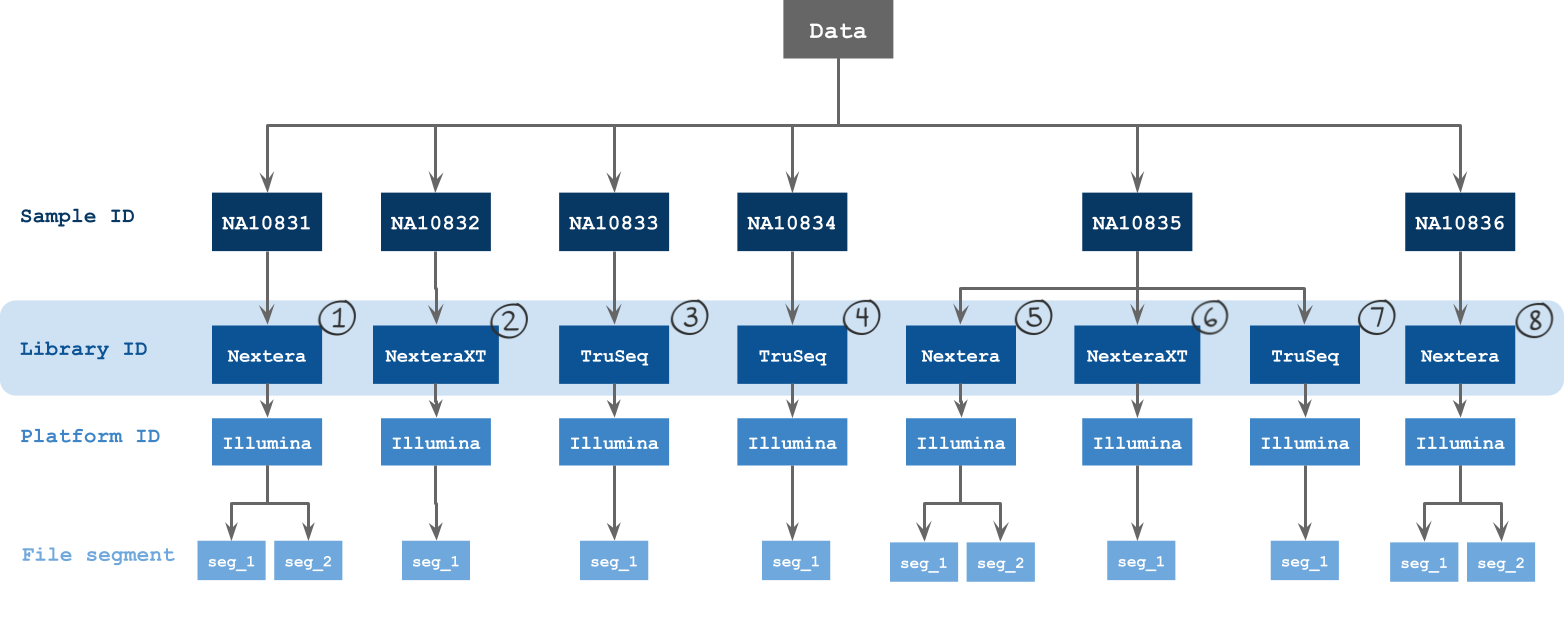
Grouping input files into batches
Group input files by their metadata or by files or run the workflow as a single task, which will group all inputs together. The optimal grouping depends on your experimental design.
For example, suppose you want to run the public Whole Genome Analysis workflow, and you have multiple FASTQ files from many samples (two paired end reads per sample, resulting in two files per sample). In this case, you might want to analyze files from each sample in batches.
Batching by File
To batch by file, select File from the Batch by drop-down menu. Batching by File runs the workflow or tool for each individual file, initiating a new child task for each input file.
Batch by file metadata
To batch by file metadata, select File metadata from the Batch by drop-down menu. Specify the metadata field by which to batch. Files are grouped by their value for a metadata field. A separate child task will be created for each different grouping.
Updated less than a minute ago
