Import Dockstore apps to the CGC
This page will explain how you can use the CGC to import an app that is available on Dockstore. Dockstore is an open platform for sharing Docker-based apps described with the Common Workflow Language (CWL), Workflow Description Language (WDL) or Nextflow, which enables bioinformaticians to share analytical tools that can be executed in a compliant execution environment, such as the CGC.
The CGC is integrated with Dockstore, which means that you can import an app from Dockstore to the CGC with a single click of a button. This process consists of two stages:
- Navigating through Dockstore and selecting an app to launch on the CGC.
- Completing the import process on the CGC and running the app.
As the CGC supports the Common Workflow Language, the CGC button is visible only for CWL apps on Dockstore. Also, please note that Dockstore is an external service and apps that are available on Dockstore are third-party apps. Therefore, Seven Bridges does not guarantee the performance of such apps on the CGC.
Finding an app on Dockstore
This step takes place on Dockstore.
- Navigate to dockstore.org in your web browser.
- In the top navigation bar click Search. The search screen opens.
- In the Search box on the left, enter the desired keyword(s).
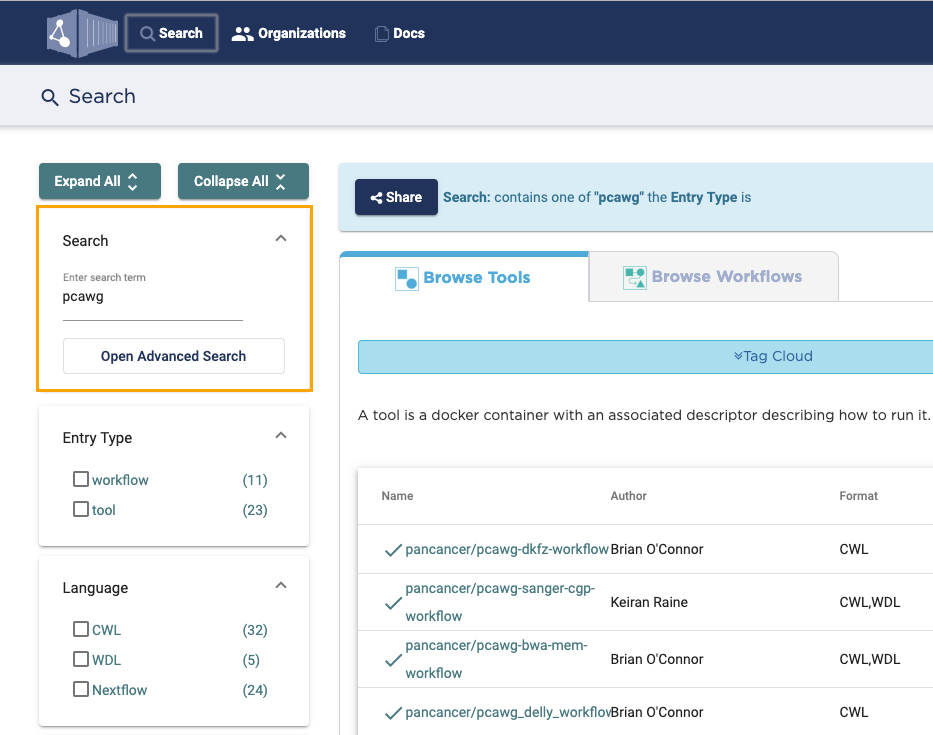
- If necessary, use additional filtering options below the Search box to narrow down the search results.
- In the search results list, click the name of the desired app. App details page opens.
- (Optional) In the Recent Versions box on the right, select a different version of the app. Latest app version is selected by default.
- In the Launch with box click CGC. This takes you to the CGC.
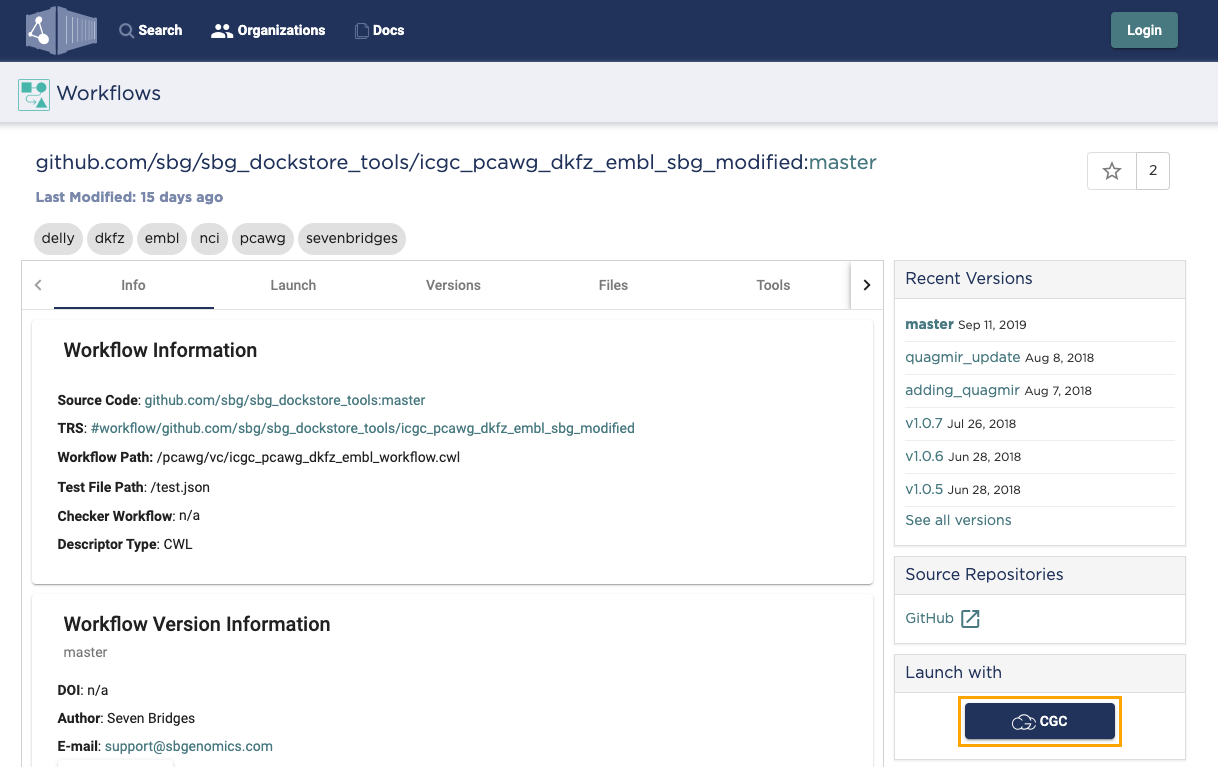
Completing the import on the CGC
This step takes place on the CGC.
After selecting Launch with > CGC on Dockstore, you are taken to the CGC and presented with the app import screen.

- Select the project in which you want to run the app.
- (Optional) Change the app URL. This is the unique URL that identifies the app on the CGC and usually does not need to be changed unless there is already an app with the same URL in the project.
- Click Import the app. This takes you to the app details page inside the selected project.
If you want to make further edits to the app before running it, click Edit in the top-right corner. Otherwise, click Run to create a draft task, set task inputs and run the app on the CGC.
Updated less than a minute ago
