Worked example of uploading SamTools Sort
Introduction
The Software Development Kit (SDK) allows you to add your tools to the CGC and use them to run analyses, as you do with tools that are already publicly available on the CGC. This is done by installing the tools in a Docker container and then describing their behavior on the CGC.
The first part of the procedure of deploying your tool to the CGC is to create a Docker container and install the tool in it. Once this is done, you need create a snapshot of the container, called an image, and push it to the CGC image registry or the official Docker image registry, Docker Hub.
The second part of the procedure is to specify the tool's behavior on the CGC, including its inputs and outputs, runtime requirements, and execution semantics. The specification is entered using the Tool Editor. This allows you to use the tool as an individual application on the CGC or interface it with other tools and create workflows.
Objective
This tutorial demonstrates how to wrap and run the sort subcommand of SAMtools on the CGC. Specifically, we shall install the bioinformatics package SAMtools into a Docker container, push it to the CGC image registry, describe the sort subcommand in the tool editor, and then use the imported sort tool in a workflow on the CGC.
Prerequisites
For this tutorial, you will need:
- An account on the CGC.
- A Linux, Mac or Windows computer with Docker installed.
Create a project
- Click Projects in the top navigation bar and choose Create a project.
- Name the project SAMtools (you can always delete this project when you've finished the tutorial).
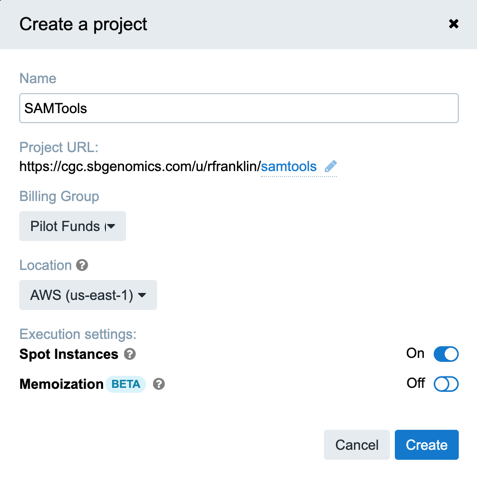
- Click Create.
You have now created a project on the CGC.
Describe each subcommand in the Tool Editor
We have created a Docker image with SAMtools inside, and pushed it to the CGC image registry. To use SAMtools on the CGC, we still need to capture its interface with the tool editor, so that it can be integrated with other CGC tools.
The tool editor treats each subcommand of a command line tool as a distinct tool. In this tutorial, we’ll wrap samtools sort, one of the tools in the samtools suite. samtools sort takes an input BAM-format file containing short DNA sequence reads and sorts it.
To describe sort:
- On the CGC, navigate to Projects > SAMtools.
- Click the Apps tab.
- Click Add App.
- Open the Create New Apptab.
- Click Create a Tool.
- Enter the name for your tool as shown below, e.g. SAMtools-sort.
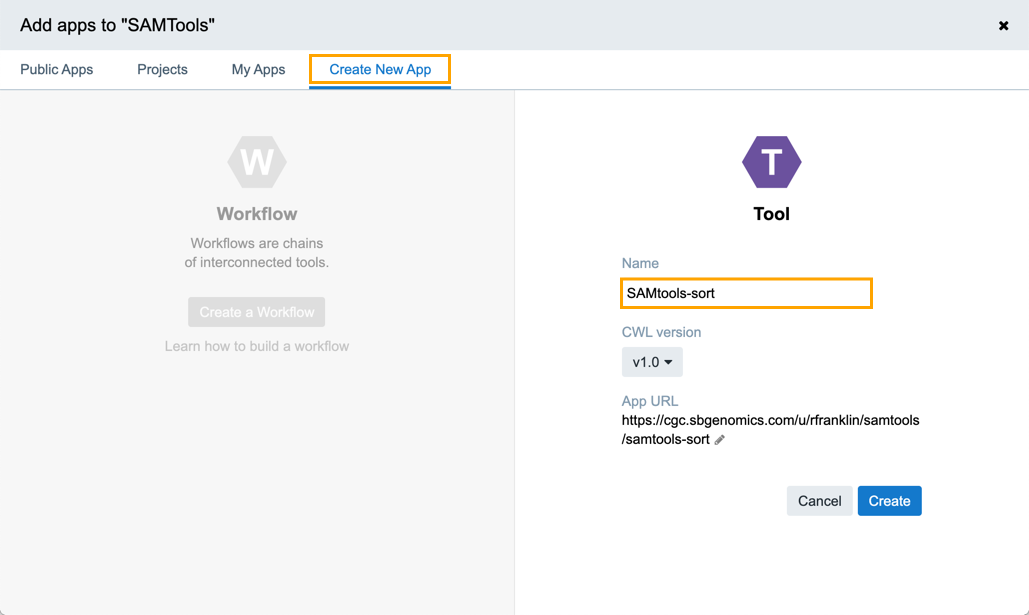
- Click Create.
The Tool Editor opens.
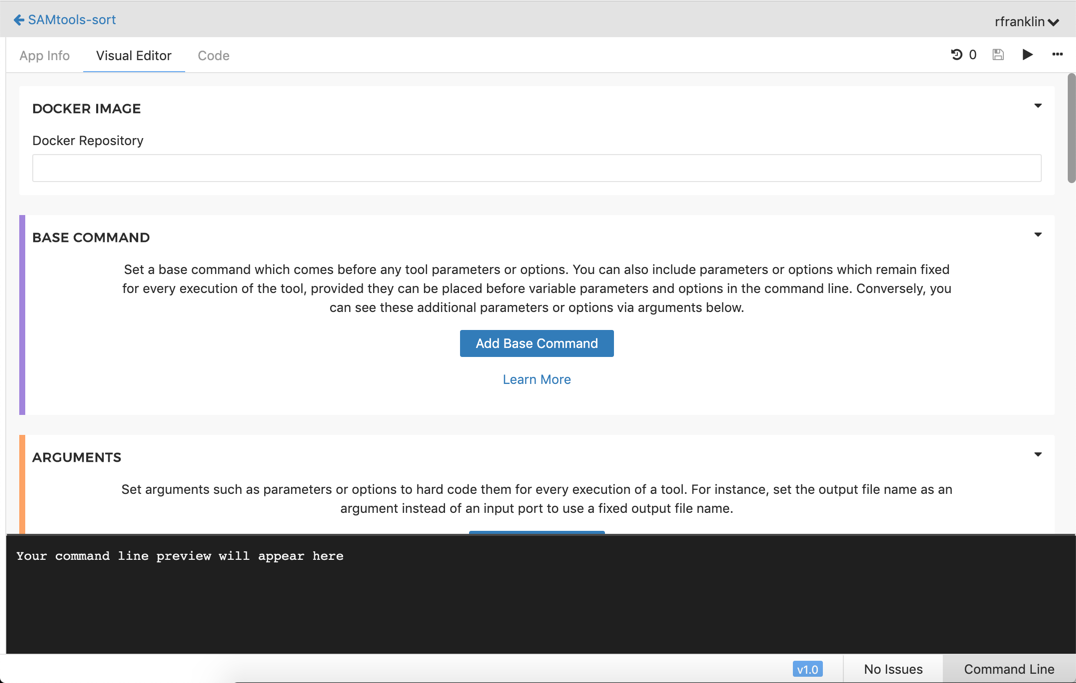
Leave it like this as we will need it later to complete the tutorial.
If your editor is significantly different than the one you see above, you're using the legacy editor. To complete the tutorial successfully, please click Switch to the new version in the banner at the top of the page to change your preferred editor.
Identify the required options for the command line
To keep the example simple, we will use the default values for most parameters and options, and aim to build a command line that looks like:
samtools sort -O bam -T tmp_ -o <sorted-bam-file.bam> <input-bam-file.bam>
The command breaks down as follows:
- samtools: the command.
- sort: the subcommand. Together, the command and subcommand form thebase command. In order for the tool to run properly, all parts of the base command that are separated by a space need to be entered in separate fields within theBase commandsection in tool editor.
- -O bam: the format of the output file. We are hard-coding this rather than allowing the user to specify it when the tool is run, so this is anargument.
- -T tmp_: the prefix to use for the temporary files. Again, we are hard-coding this rather than allowing the user to specify it when the tool is run, so this is anargument.
- -o <sorted-bam-file.bam>: the name of the output file to generate. We want to allow the user to specify the name of the output file as an input to the command, so this is an input port. Note that the file that is generated will be anoutput port.
- <input-bam-file.bam>: the BAM file to be sorted. We want to allow the user to specify this file as an input to the command, so this is an input port.
Alternatively, we could use a dynamic expression to derive the output file name from the input file name. For example, we could set the output file name to<input-bam-file>_sorted.bamwhere<input-bam-file>is the first part of the input file name. In this case, when the tool is run, the user will not need to specify a value for the output filename. So the output filename parameter is no longer an input port (specified when the tool is run) but an argument containing a dynamic expression (either fixed, or derived automatically from other information). We won’t use dynamic expressions in this tutorial, but in the next tutorial, we’ll see how to modify this example to use a dynamic expression.
Create the Docker image
Note: A Docker image in the image repository can be accessed by anyone who knows the path and name. So you should avoid including any sensitive data in the image or set the repository privacy settings to Private.
Make sure Docker is running
- Mac OS 10.10.3 Yosemite or newer: run Docker Desktop for Mac and start a terminal of your choice.
- Mac OS 10.8 Mountain Lion or newer: use Docker Toolbox.
- Windows 10: run Docker for Windows and start a terminal of your choice.
- Windows 7 or 8: run Docker Quickstart Terminal to start Docker Machine.
- Linux: no action required.
Open a terminal window on your computer and enter the following command:
docker run -ti ubuntu:16.04
This creates a Docker container from the ubuntu base image. Here, we are using a minimal ubuntu base image as that is suitable for samtools, but you can start with any image that is suitable for the tools you want to use.
The terminal prompt changes to root@ where is a set of 12 alphanumeric characters that represent the unique id for the Docker container you are creating. Make a note of as you will need it shortly.
Load the container with the tools you need. In this case, you need to enter the following commands to download and build samtools.
# Update the package index inside the container
apt-get update
# Install the tools we need to download and compile SamTools
apt-get install wget build-essential zlib1g-dev libncurses5-dev liblzma-dev libbz2-dev
# Download the SAMtools source code (version 1.6 or a later version of you prefer)
wget https://github.com/samtools/samtools/releases/download/1.6/samtools-1.6.tar.bz2
# Unpack the archive
tar jxf samtools-1.6.tar.bz2
# Go into the directory containing the unpacked Samtools source code
cd samtools-1.6
# Compile the code
make
# Install the resulting binaries
make install
Test that the samtools executable has been installed and built successfully. Enter the following command to verify that the version information for samtools is displayed.
samtools --version
If everything is installed properly, the output should look like this:
samtools 1.6
Using htslib 1.6
Copyright (C) 2017 Genome Research Ltd.
Exit the container (remember to make a note of the container id).
exit
Save and upload the Docker image
To save the container as an image in the CGC image registry, first log in to the registry. In the terminal window, enter:
docker login cgc-images.sbgenomics.com
When prompted for a username, enter your CGC username. When prompted for a password, enter your CGC authentication token, not your CGC login password.
You will see a message saying the login has succeeded, then you will be returned to the terminal prompt. Note that this login times out after a while, so if you don’t access the CGC image registry promptly, you may need to log in again in order to do so.
Commit the image to the repository as follows:
docker commit <containerid> cgc-images.sbgenomics.com/<username>/samtools:v1
In this command, <containerid> is the ID of the container you made a note of above, <username> is your CGC username. In this example, we have called the image samtools, and have tagged it as v1. If the commit is successful, you will see a message similar to this:
sha256:4dcd3c6911776ba0417e322dd40d0d4881e1806f9b3027516888798b21b8203f
Push the image to the image registry:
docker push cgc-images.sbgenomics.com/<username>/samtools:v1
where <username> is your CGC username. Please wait for the procedure to complete.
If the push is successful, you will see several messages, ending with a message similar to this:
v1: digest: sha256:64a47b5dcdb95a4b6184e880365694b40e1cd85e4151074a11ba1f37c8b56f1f size: 1570
If you want to know more about Docker commands, you will find a list of common Docker commands here.
Specify the Docker image containing the tool
Go back to your SAMtools-sort project on the CGC. In the Docker Image section of the tool editor, set Docker repository to cgc-images.sbgenomics.com/<username>/samtools:v1 where <username> is your CGC username.
Specify the base command
- In the Base Command section of the tool editor, click Add Base Command.
- Enter samtools.
- Under the text field click + Add Base Command.
- Enter sort in the blank field.
Click Command Line at the bottom right to open a preview pane showing a preview of the command we are building up. You should see samtools sort in the preview pane.
Specify the arguments
We need to specify the output file format as a fixed argument (the string is -O bam).
In the Arguments section of the tool editor, click Add an Argument. An argument is added and the object inspector opens on the right hand side showing the properties of the argument.
In the object inspector:
- Leave Use command line binding selected
- Set Prefix to -O
- Set Value to bam
- Leave Separate value and prefix selected (the syntax requires a space between the prefix and the expression)
- Leave Position set to 0 (as long as this argument is after the base command at the beginning of the command line and before the input file at the end of the command line, the actual position relative to the other items on the command line doesn’t matter).
We also need to specify the temporary file prefix as a fixed argument (the string is -T tmp_).
In the bottom-left part of the Arguments section, click + Add an Argument. Then, in the object inspector on the right:
- Leave Use command line binding selected.
- Set Prefix to -T
- Set Value to tmp_
- Leave Separate Value and Prefix selected (the syntax requires a space between the prefix and the expression)
- Leave Position set to 0 (as long as this argument is after the base command at the beginning of the command line and before the input file at the end of the command line, the actual position relative to the other items on the command line doesn’t matter).
In the preview pane you should see samtools sort -O bam -T tmp_.
Specify the input ports
We need to specify the name of the output file as an input port (the string is -o .bam).
In the Input ports section of the tool editor, click Add an Input. An input port is added, with a default name of input, and the object inspector opens on the right hand side showing the properties of the input.
In the object inspector on the right:
- Select Required. This will be a mandatory input.
- Set ID to sorted_file_name.
- Set Type to string.
- Leave Allow array as well as single item unselected.
- Select Include in the command line.
- Leave Value Transform blank. This is where we could insert a dynamic expression to derive the name as a function of the input file name if we wanted to. Because we have left this blank, the user of the tool will be asked to specify a value when the tool executes.
- Set Prefix to -o.
- Leave Separate value and prefix selected.
- Set Position to 0 (as long as this part of the command is after the base command at the beginning of the command line and before the input file at the end of the command line, the actual position relative to the other items on the command line doesn’t matter).
- Expand the Description drop-down, and set Label to Sorted file name. Optionally, add a more detailed description of the input port in the Description text box. When the tool is placed in a workflow, the value from the Label field is displayed against the output port (if not supplied, the ID is used instead).
In the preview pane, you should see samtools sort -O bam -T tmp_ -o sorted_file_name-string-value.
We also need to specify the input file as an input port. In the bottom-right part of the Input ports section, click + Add an Input.
In the object inspector on the right:
- Select Required.
- Set ID to input_bam_file.
- Set Type to File.
- Leave Value Transform blank.
- Leave Prefix blank.
- Leave Separate Value and Prefix selected.
- Set Position to 1 (this must appear in the command line after the other arguments and inputs).
- Scroll down to the Description section, expand the drop-down, and set Label to Input BAM file. Optionally, add a more detailed description of the input port in the Description text box.
- Set File type(s) to BAM (only valid for CWL v1.x workflows). When the tool is placed in a workflow, the value from the Label field is displayed against the input port (if not supplied, the ID is used instead). For CWL v1.x tools only, File type(s) allows the workflow editor to check that output nodes are connected to input nodes of the correct type.
In the preview pane, you should see samtools sort -O bam -T tmp -o sorted_file_name-string-value /path/to/input.ext.
Specify the output port
Now we need to specify the output file as an output port. Note that we have already set the name of the output file as an input. But we also need to specify an output port for the file in order to retrieve the output. In the Output ports section of the tool editor, click Add an Output. An output port is added and the object inspector opens on the right hand side showing the properties of the output.
In the object inspector:
- Select Required.
- Set ID to sorted_bam_file.
- Set Type to File.
- Set Glob to *.bam. This means that any file that matches this filter will be reported as an output of the tool. We could use a dynamic expression instead to specify only files that match the specified output file name, but this simpler option will be enough for now.
- Scroll down to the Description drop-down, expand it, and set Label to Output BAM file. Optionally, add a more detailed description of the output port in the Description text box.
- Set File type(s) to BAM (only valid for CWL v1.x workflows). When the tool is placed in a workflow, the value from the Label field is displayed against the output port (if not supplied, the ID is used instead) and, for CWL V1.x tools only, File type(s) allows the workflow editor to check that output nodes are connected to input nodes of the correct type.
Save the tool
- Click the Save icon in the top-right corner to save the tool description.
- (Optional) Add a short revision note describing the changes you have made.
- Click Save.
Test the tool
We are going to use a typical BAM file from the 1000 Genomes project to test the tool on the CGC, so first you need to copy it to your project.
- On the CGC, select Data > Public Test Files.
- Enter NA12878.ga2.exome.maq.raw.bam in the search box. Note that the task will take around 75 minutes to execute with this input file. If you want to do only a quick test of the tool, you could use a smaller input file, as this will return a result in a few minutes. If so, search for G26234.HCC1187_1M.aligned.bam instead.
- Select the file then click Copy, and specify the project where your newly-created tool is located.
- Navigate back to the project where your samtools-sort tool is located.
- Click the Apps tab.
- Click Run next to samtools-sort.
- Next to the Input BAM file input port click Select File(s), and select NA12878.ga2.exome.maq.raw.bam. If you opted for using the smaller file in step 2 above, select G26234.HCC1187_1M.aligned.bam instead.
- Click Save selection.
- In the Sorted file name field enter sorted_bam_file.bam.
- Click Run.
This analysis will take around 75 minutes to run (or less if you are using the smaller input file), and you will receive an email when it is completed.
View the results
When you receive the notification email, click the link in the email to view the results. You should see that the task was successful and that a single output file named sorted_bam_file.bam was created,
Updated less than a minute ago
