Import data from ICDC
About ICDC
Integrated Canine Data Commons (ICDC), a cloud-based repository of canine cancer data and was established to further research on human cancers by enabling comparative analysis with canine cancer. The data in the ICDC is sourced from multiple different programs and projects; all focused on canine subjects. The data is harmonized into an integrated data model and then made available to the research community.
Data that is available for import on the CGC is now instantly aligned with ICDC data updates. With the power of global standards for data sharing all data that ICDC provides access to can be imported and used on the CGC platform.
The process of importing files from ICDC to the Cancer Genomics Cloud (CGC) is simple and can be done by a push of a button on the ICDC portal.
Users that are already familiar with the old process can use the following two stages:
- Downloading a manifest file from the ICDC website.
- Importing files to the CGC based on the downloaded manifest file.
Import files from ICDC to the CGC
To import ICDC files you must first find the data of interest on the ICDC portal:
- Open the ICDC website.
- Click Explore in the main navigation bar at the top of the page.
- In the filters section on the left, select criteria to narrow down the cases list.
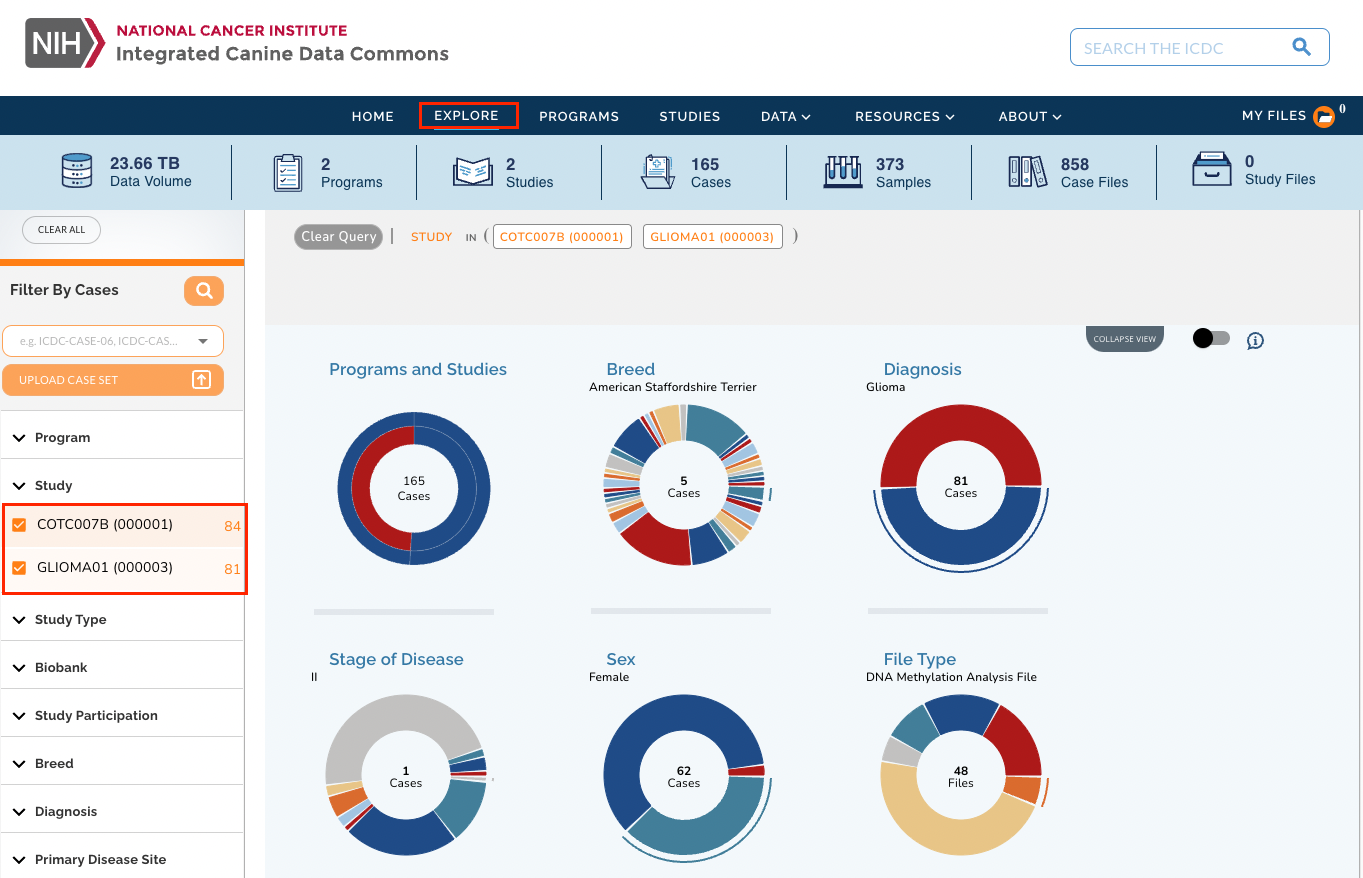
- Scroll down to see the list of cases that match the criteria.
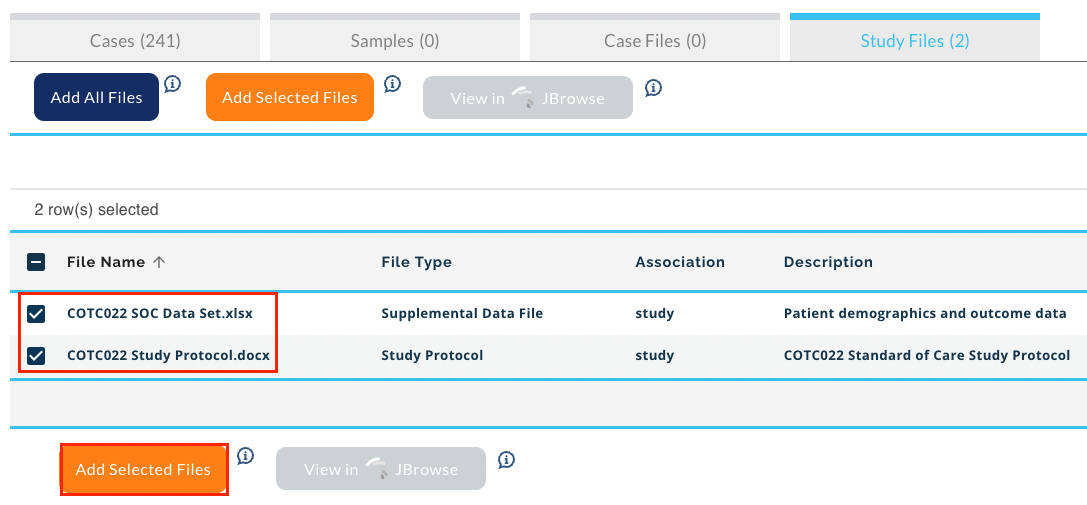
- Click Study Files.
- Select the files and click Add Selected Files.
- Click MY FILES in the top right corner of the page. The information about the number of selected files is also shown.
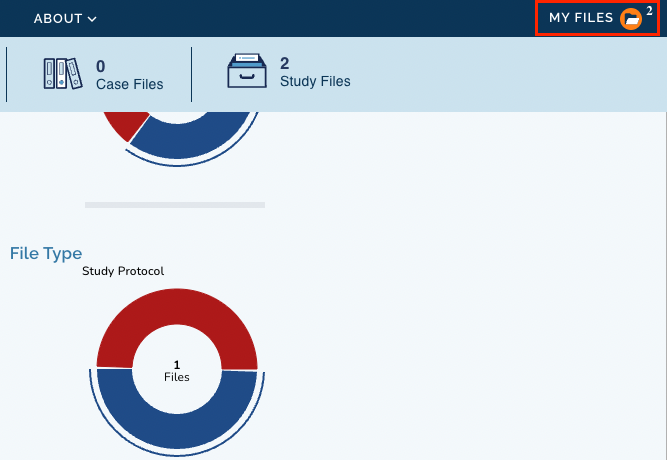
-
Select the files which you want to import on CGC.

-
Click Available Export Options and choose "Export to Cancer Genomics Cloud". You will then land on CGC where you can continue with the procedure of importing files.
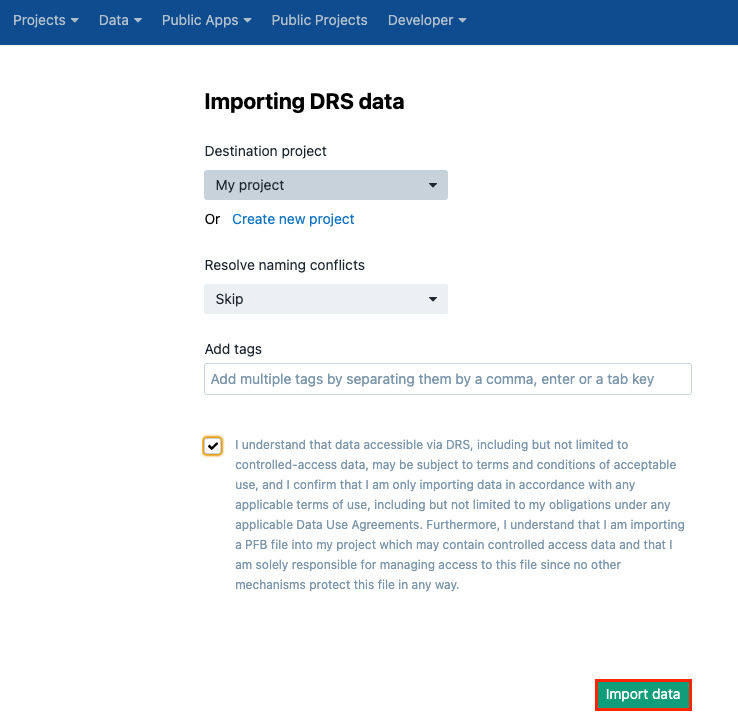
- Choose the destination project and set the rule for resolving naming conflicts.
- (Optional) Specify tags for these files.
- Tick the box to comply with the rules for using these files.
- Click Import data.
The files are now imported to your project.
Import files using a manifest file
An alternative way of importing files from the ICDC portal is by using the manifest file. Follow these steps:
- After selecting files you want to import (see above for more information as the procedure is the same) and adding files to the My Files section, click Download File Manifest.
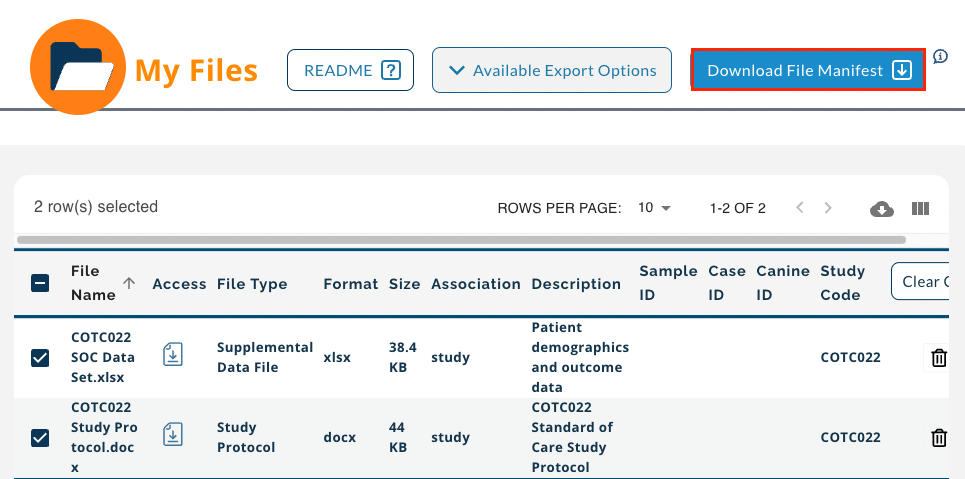
-
In the popup that is displayed next, click Continue to Download File Manifest.
-
Save the manifest to your computer. The rest of the steps are done on the CGC.
-
Access the project you want to import the files to.
-
Click the Files tab.
-
Click Add files and choose "GA4GH Data Repository Service (DRS)".
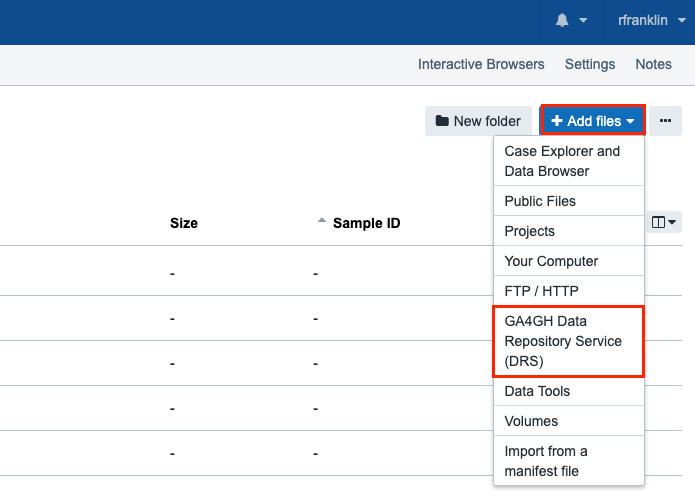
-
Next, click From a manifest file.
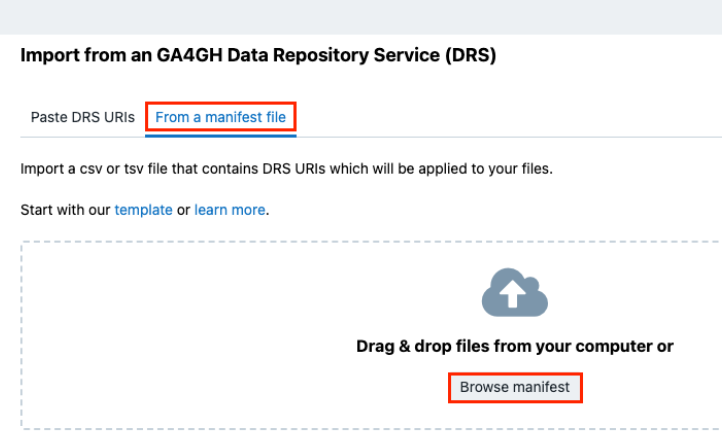
-
Click Browse manifest and locate the manifest file you have previously downloaded from the ICDC Portal.
-
Click Submit.
The files are now imported to your project.
Metadata
Metadata assigned to each ICDC file is now managed by ICDC portal and is set during the import process. If you want to get more metadata from ICDC, you can use the ICDC API, which provides easy and open access to ICDC metadata. One of the ways to get ICDC metadata on the CGC is to use Data Cruncher to query the ICDC API.
Reference files
The Public Reference Files repository on the CGC contains two ICDC-related files that are ready to be used in e.g. RNA-seq alignment workflows with ICDC data provided as inputs. Those files are:
Canis_familiaris.CanFam3.1.dna.toplevel.fa(reference genome)Canis_familiaris.CanFam3.1.98.gtf(gene annotation file)
Updated almost 2 years ago
