Data Cruncher Quickstart
Overview
Data Cruncher is an interactive analysis tool on the CGC for exploring and mining data using Jupyter notebooks.
Objective
The aim of this guide is to show you how to create and run your first analysis in Data Cruncher, using a real-life example of filtering VCF files based on the alternative allele read depth (AD) and the read depth (DP) ratio.
Procedure
- [ 1 ] Access Data Cruncher from a project on the Platform.
- [ 2 ] Create and set up an analysis.
- [ 3 ] Enter and execute code within the analysis to get results.
Prerequisites
- You need to download this sample generic VCF file and upload it to the project on the CGC in which you want to execute the analysis.
- You need execute permissions in the project to be able to run the analysis.
[ 1 ] Access Data Cruncher
- Open the project on the CGC that contains the uploaded VCF file.
- From the project's dashboard, click the Interactive Analysis tab.
The list of available interactive analysis tools opens. - On the Data Cruncher card click Open.

This takes you to the Data Cruncher home page.
[ 2 ] Create and set up your first analysis
If there are no previous analyses, the main Data Cruncher screen will be blank.
- Click Create your first analysis.
The Create new analysis wizard is displayed. - On the first screen, enter VCF Filtering in the Analysis name field.
- Click Next.
- Select the instance for the analysis.
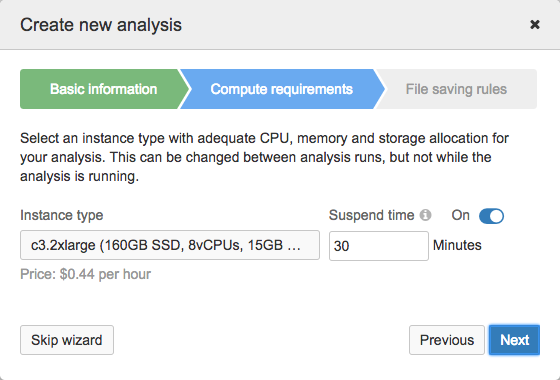
The Instance type list displays available instances along with their disk size, number of vCPUs and memory (shown in brackets). The default instance is c3.2xlarge that has 160 GB of SSD storage, 8 vCPUs and 15 GB of RAM.
Suspend time is the period of analysis inactivity after which the instance is stopped automatically. Inactivity implies that:
- No files have been modified or created under the Files tab (in the
/sbgenomics/workspacedirectory if you are using the Terminal). - There are no running kernels.
Apart from stopping the instance, this also includes stopping the analysis and saving all files that meet the criteria for automatic saving or have been selected to be saved as project files. Files that do not meet the criteria and are not manually saved to the project will be lost. Minimum suspend time is 15 minutes.
- Click Next.
- Define the automatic saving criteria:
- Ignore the following file types - Files that have the listed extensions will never be automatically saved when the analysis is stopped. We will keep the suggested ignored file types,
.log, .zip. - Ignore files larger than - Files bigger than the specified size will not be automatically saved when the analysis is stopped.
- Click Start the analysis.
The CGC will start acquiring an adequate instance for your analysis, which may take a few minutes. Once an instance is ready, you will be notified.
[ 3 ] Start the analysis
Once the CGC has acquired an instance for your analysis, you can open the editor and run your analysis.
- Click Open in editor.
The editor opens in a new window. - On the welcome dialog, click Notebook.
- Enter the notebook details:
- File Name - Name the notebook VCF_Filtering.
- Kernel - This is the “computational engine” that executes the code contained in a notebook. Select Python 2.
- Click Create.
Your notebook is now ready. You can start entering the code and the additional text. - In the first cell paste the following code:
import pandas as pd
- Click
 on the toolbar. This executes the current cell and creates a new one below.
on the toolbar. This executes the current cell and creates a new one below. - Click Code on the toolbar and select Markdown from the dropdown list. This changes the cell type to Markdown, so we can add a title.
- Paste the following text into the cell:
# Constants and Functions
- Click
 .
. - Paste the following code into the next cell:
vcf_column_names = ["CHROM", "POS", "ID", "REF", "ALT", "QUAL", "FILTER", "INFO", "FORMAT", "NORMAL", "TUMOR"]
def read_vcf_df(vcf_path, vcf_column_names):
return pd.read_csv(vcf_path, comment="#", header=None, names = vcf_column_names , sep="\t")
def ad_dp_calc(x):
ref = x[0]
alts = x[1].split(',')
gt = x[2].split(':')
if len(gt) == 1:
return 0
dp = float(gt[0].replace(",","."))
a = float(gt[4].replace(",","."))
c = float(gt[5].replace(",","."))
g = float(gt[6].replace(",","."))
t = float(gt[7].replace(",","."))
ad = 0
for alt in alts:
if alt == "A":
ad += a
elif alt == "C":
ad += c
elif alt == "G":
ad += g
elif alt == "T":
ad += t
return float(ad) / (dp + len(alts))
def read_vcf_header(file_name):
header = ""
with open(file_name) as f:
for line in f:
if line[0] == "#":
header += line
else:
break
return header
- Click
 .
. - Change the type of the blank cell to markdown and paste the following text:
# AD/DP VCF Filtering Description
We are **filtering VCF files** based on the alternative allele read depth (**AD**) and the read depth (**DP**) ratio. We are discarding all the variants that don't pass the criteria of **AD/DP > 0.15**.
- Click
 .
. - Change the type of the blank cell to markdown and paste the following text:
# VCF Filtering
- Click
 .
. - Paste the following code:
vcf_name = "PATH_TO_VCF_FILE"
vcf = read_vcf_df(vcf_name, vcf_column_names)
title_name = "Distribution of AD/DP Ratios for FPs SNVs GRAL"
vcf["AD_DP"] = vcf[["REF","ALT","TUMOR"]].apply(lambda x: ad_dp_calc(x), axis=1)
vcf = vcf[vcf["AD_DP"] >= 0.15 ]
del vcf["AD_DP"]
header = read_vcf_header(vcf_name)
with open("REALIGNED_TUMOR.bam.filtered.vcf","w") as f:
f.write(header)
vcf.to_csv(f, sep="\t", header=False, index=False )
On line 1 above, there is a placeholder named PATH_TO_VCF_FILE that we need to replace with the actual path to the uploaded file:
a. Open the Project Files tab.
b. Click TEST.bam.vcf. This will copy the file path to the clipboard.
c. Paste the copied path instead of PATH_TO_VCF_FILE on line 1 of the code block above, making sure to keep the quotation marks.
17. Click  .
.
The analysis now executes. A new file named REALIGNED_TUMOR.bam.filtered.vcf is created and you can see it under the Files tab in the analysis editor.
Updated less than a minute ago
