API Batch tutorial
On this page:
- Overview
- Objective
- Prerequisites
- Procedure
- Step 1: Find your billing group
- Step 2: Create a project
- Step 3: Add data files to your project
- 3a: Find your files
- 3b: Copy files to a project
- Step 4: Add reference files to your project
- 4a: Find reference files
- 4b: Copy reference files to your project
- Step 5: Add a public workflow to your project
- 5a: Find a public workflow
- 5b: Copy a public app into a project
- 5c: Modify a workflow on the visual interface
- 5d: Find an app in your project via the API
- Step 6: Create a draft task
- Step 7: Run a task
- Step 8: Get task outputs
- 8a: List the child tasks
- 8b: Obtain the outputs of a child task
This tutorial introduces you to performing a batch analysis using the API.
Batching allows you to run identical analyses on different data, by entering multiple input files and grouping them with specified metadata criteria. For instance, you can group input files by File, Sample, Library, Platform unit, or File segment. By using Batch Input, you can process multiple datasets with a single workflow containing the same parameter settings without having to set up the workflow multiple times. Batching creates one parent task containing multiple child tasks: one for each group of files.
In this tutorial, we'll run a batch analysis in which we align reads based on their Sample metadata.
You will need an account on the CGC in order to obtain your authentication token. Almost all API requests require your CGC authentication token. This acts as a security measure regulating your access to your projects. Learn more about obtaining your authentication token.
##Procedure
We'll use the API to create a project and populating it with files. Then, we'll use the visual interface to modify one of the RNA sequencing workflows, RNA-seq Alignment STAR, to carry out the analysis. At this point, we will use the API to specify the inputs and set the batch criteria to batch by sample. Finally, we'll examine our results.
All necessary tools and data are available on the CGC.
We will state the HTTP requests for each API call used in the procedure. But you could also write a script that makes these calls using Seven Bridges Python or R client libraries.
##Step 1: Find your billing group
To start an analysis, we must first create a project. To do this, we need to obtain following information:
- Your CGC authentication token
- A billing group ID
Your authentication token acts as a security measure so only you can access your projects and resources on the CGC. The billing group ID designates which funding resource to charge for the analyses you run in the project you're about to create. Learn more about billing groups on the CGC.
Use the API request to list your billing groups, as shown in the HTTP request below. Be sure to substitute your authentication token for X-SBG-Auth-Token.
GET /v2/billing/groups HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
This request returns a list of the billing groups you are part of, as shown below:
{
"href": "https://cgc-api.sbgenomics.com/v2/billing/groups/",
"items":
{
"id": "ec1dc1e3-12a3-4b56-789c-e3f2dca0c6f7",
"href": "https://cgc-api.sbgenomics.com/v2/billing/groups/ec1dc1e3-12a3-4b56-789c-e3f2dca0c6f7",
"name": "My Funds (rfranklin)"
}
"links": []
}
Copy the value for id (in this case ec1dc1e3-12a3-4b56-789c-e3f2dca0c6f7) to your clipboard. We will use this in the next step when creating a project.
Projects on the CGC serve as the containers for the data, analytical tools, results, and team of collaborators for a distinct scientific investigation.
To create a project, make the API request to create a project, as shown in the HTTP request shown below. Be sure to paste in your authentication token for the X-SBG-Auth-Token key.
This request also requires a request body. Provide a name for your project and an optional description. Here, you should also paste in the billing_group id you obtained in the previous step.
POST /v2/projects HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
content-type: application/json
{
"name":"Batch tutorial",
"description":"project for batching by sample via the APi",
"billing_group":"ec1dc1e3-12a3-4b56-789c-e3f2dca0c6f7"
}
You'll see a response body, as shown below, containing the name of your project, its URL (href), your project id, and your project's billing_group.
Note down the project id. We will use this throughout the tutorial to designate our project. The project id consists of two parts: your username followed by your project's short name.
{
"href": "https://cgc-api.sbgenomics.com/v2/projects/rfranklin/batch-tutorial",
"id": "rfranklin/batch-tutorial",
"name": "Batch-tutorial",
"type": "v2",
"description": "project for batching by sample via the API",
"tags": [],
"billing_group": "ec1dc1e3-12a3-4b56-789c-e3f2dca0c6f7"
}
Now that you've successfully created a project, we can add data to the project for analysis.
##Step 3: Add data files to your project
In this tutorial, we'll analyze data that is hosted in the Cancer Cell Line Encyclopedia (CCLE) public project on the CGC.
Public projects are repositories for examples of specific analyses as well as the associated data and tools you need to replicate these analyses on the CGC.
For this analysis, we want to use data files contained within the CCLE project.
To find BAM files that contain RNA sequencing data within CCLE, we will make the API request to list all files in a project, as shown below.
We'll need to pass along two query parameters to locate the files. First, we have to specify the project containing the files. In this case, the CCLE public project is specified by the id of sevenbridges/cancer-cell-line-encyclopedia-ccle-1. Following the path, you can pass this query parameter using project=sevenbridges/cancer-cell-line-encyclopedia-ccle-1.
Then, we want to find BAM files with an experimental strategy of RNA-Seq. It is possible to filter by metadata fields to retrieve files with certain properties. In this tutorial, however, we already know that we want to find the following three files:
- G30630.VM-CUB1.3.bam
- G30603.TUHR4TKB.1.bam
- G28034.MDA-MB-361.1.bam
Since we know the files' names, we can filter the returned files by name, using the name query parameter. We can append this query parameter to the query parameter project using an ampersand (&).
You can search for multiple files by name with the same API request, by including the field name multiple times. When filtering on any resource, including the same field several times with different filtering criteria results in an implicit OR operation for that field and the different criteria, so including the name field three times for three file names will find files matching any one of the file names.
The HTTP request to find files in the CCLE project matching our chosen file names is shown below:
GET /v2/files?project=sevenbridges/cancer-cell-line-encyclopedia-ccle-1&name=G30630.VM-CUB1.3.bam&name=G30603.TUHR4TKB.1.bam&name=G28034.MDA-MB-361.1.bam HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
In the response returned, you will see a list of files along with their id, name, and the project to which they belong, as shown below.
{
"href": "https://cgc-api.sbgenomics.com/v2/files?offset=0&name=G30630.VM-CUB1.3.bam&name=G30603.TUHR4TKB.1.bam&name=G28034.MDA-MB-361.1.bam&limit=3&project=sevenbridges/cancer-cell-line-encyclopedia-ccle-1",
"items": [
{
"href": "https://cgc-api.sbgenomics.com/v2/files/57da918fe4b002eed2cb10eb",
"id": "57da918fe4b002eed2cb10eb",
"name": "G30603.TUHR4TKB.1.bam",
"project": "sevenbridges/cancer-cell-line-encyclopedia-ccle-1"
},
{
"href": "https://cgc-api.sbgenomics.com/v2/files/57da918fe4b002eed2cb0915",
"id": "57da918fe4b002eed2cb0915",
"name": "G28034.MDA-MB-361.1.bam",
"project": "sevenbridges/cancer-cell-line-encyclopedia-ccle-1"
},
{
"href": "https://cgc-api.sbgenomics.com/v2/files/57da918fe4b002eed2cb0fe7",
"id": "57da918fe4b002eed2cb0fe7",
"name": "G30630.VM-CUB1.3.bam",
"project": "sevenbridges/cancer-cell-line-encyclopedia-ccle-1"
}
],
"links": []
}
Copy the id for each of the three files to your clipboard. We will use this in the next step when copying these files into our project.
###3b: Copy files to a project
To copy files into a project, make the API request to batch copy files, as shown below.
In the body of the request, you can designate the target project for the copied files. As shown in the example request below, supply a project id consisting of your username followed by the project's short name for the project key.
We also want to pass along the file ids you obtained in the step above in the body of the request. We can input the ids as a list of values for the file_ids key, as shown below.
POST /v2/action/files/copy HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
{
"project":"rfranklin/batch-tutorial",
"file_ids":[
"567890abce4b002eed2cb10eb",
"567890abc4b002eed2cb0915",
"567890abc4b002eed2cb0fe7"
]
}
The response body, as shown below, will indicate if your request was successful. The response contains the original ids of your copied files and the status of the response.
The response body also contains two other fields: new_file_id and new_file_name. These indicate the new id and the new name assigned to the copy of the file within your project. You can use this id in future API requests to refer to the copy of the file within your project as opposed to the original file.
{
"567890abc4b002eed2cb10eb": {
"status": "OK",
"new_file_id": "567890abc4b002eed2cbe64d",
"new_file_name": "G30603.TUHR4TKB.1.bam"
},
"567890abc4b002eed2cb0915": {
"status": "OK",
"new_file_id": "567890abc4b002eed2cbe64f",
"new_file_name": "G28034.MDA-MB-361.1.bam"
},
"567890abc4b002eed2cb0fe7": {
"status": "OK",
"new_file_id": "567890abc4b002eed2cbe651",
"new_file_name": "G30630.VM-CUB1.3.bam"
}
}
The files have been successfully copied to your project.
##Step 4: Add reference files to your project
Many bioinformatics tools require certain data, such as reference genomes or annotation files, to execute properly. The CGC maintains a collection of the latest and most frequently used reference genomes and annotation files in the Public Reference Files repository. The RNA-seq Alignment STAR workflow uses a reference genome and an annotation files to align reads. We'll need to have these reference files in our project to be able to use them while setting up our task.
For this analysis, we need to supply the workflow with the following two reference files:
- HG19_Broad_variant.fasta
- Homo_sapiens.GRCh37.75.gtf
See the table below for more information about each file.
| API key | Input files | File type |
|---|---|---|
genomeFastaFiles | HG19_Broad_variant.fasta | FASTA is a reference genome file which we will use for the alignment of the FASTQ files. |
sjdbGTFfile | Homo_sapiens.GRCh37.75.gtf | GTF is an annotation file containing information about gene structure. |
To find the reference files above in the Public Reference Files repository, we will make the API request to list all files in a project, as shown below.
This process is similar to finding the data files above. In this case, the Public Reference Files repository is specified in the same way as a project on the CGC by an id of admin/sbg-public-data. Following the path, you can pass this query parameter using project=admin/sbg-public-data.
As above, we want to filter the results by the name parameter to find HG19_Broad_variant.fasta and Homo_sapiens.GRCh37.75.gtf.
The entire HTTP request is shown below:
GET /v2/files?project=admin/sbg-public-data&name=HG19_Broad_variant.fasta&name=Homo_sapiens.GRCh37.75.gtf HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
In the response, you will see information about each file as well as the file's id. Copy each id (for example, 5772b6cd507c1752674486d8) to your clipboard. We will use these ids in the next step.
{
"href": "https://cgc-api.sbgenomics.com/v2/files?offset=0&name=HG19_Broad_variant.fasta&name=Homo_sapiens.GRCh37.75.gtf&limit=2&project=admin/sbg-public-data",
"items": [
{
"href": "https://cgc-api.sbgenomics.com/v2/files/5772b6c4507c1752674486cd",
"id": "5772b6c4507c1752674486cd",
"name": "Homo_sapiens.GRCh37.75.gtf",
"project": "admin/sbg-public-data"
},
{
"href": "https://cgc-api.sbgenomics.com/v2/files/5772b6c1507c1752674486c9",
"id": "5772b6c1507c1752674486c9",
"name": "HG19_Broad_variant.fasta",
"project": "admin/sbg-public-data"
}
],
"links": []
}
Pro-tip:
To display only the
idandnamefields in the response, you can specifyfieldsas a query parameter by usingfields=id,name.
###4b: Copy reference files to your project
To copy files into a project, make the API request to batch copy files, as shown below. This is the same method we used to copy our data files into our project.
In the body of the request, you can specify the target project for the copied files, such as rfranklin/batch-tutorial, as a value for the project key.
We also want to pass along the file ids you obtained in the step above in the body of the request. We can input the ids as a list of values for the file_ids key, as shown below.
POST /v2/action/files/copy HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
{
"project":"rfranklin/batch-tutorial",
"file_ids": [
"567890abc07c1752674486cd",
"567890abc07c1752674486c9"
]
}
The response body contains the new_file_id for each of the copied reference files. These indicate the new id assigned to the copy of the file within your project. You can use this id in future API requests to refer to the copy of the file within your project as opposed to the original file
{
"567890abc07c1752674486cd": {
"status": "OK",
"new_file_id": "567890abc4b002eed2cbf489",
"new_file_name": "Homo_sapiens.GRCh37.75.gtf"
},
"567890abc07c1752674486c9": {
"status": "OK",
"new_file_id": "567890abc4b002eed2cbf48b",
"new_file_name": "HG19_Broad_variant.fasta"
}
}
We have populated our project with the requisite data and reference files. Now, we can add a workflow to our project.
##Step 5: Add a public workflow to your project
We need a workflow to analyze our data. We'll start with a publicly available workflow from the Public Apps repository, RNA-seq Alignment - STAR. However, the workflow takes FASTQ inputs. Since we added BAM files from CCLE above, we'll need to modify the workflow to accept input BAM files and convert them to the FASTQ format.
To find a public workflow on the CGC, make the API request to list all apps (i.e. tools and workflows) available to you, as shown below.
You can filter for public workflows by adding the parameter visibility=public. Since this will return many results, we want to see as many results as we can on one page. To set the pagination, we use the query parameter limit=100 to display 100 results per page. The maximum allowable limit per page is 100.
GET /v2/apps?visibility=public&limit=100 HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
This query returns the response below. For brevity, we have omitted part some of the returned apps.
{
"href": "https://cgc-api.sbgenomics.com/v2/apps?visibility=public&offset=0&limit=100",
"items": [
{
"href": "https://cgc-api.sbgenomics.com/v2/apps/admin/sbg-public-data/sbg-split-bed/3",
"id": "admin/sbg-public-data/sbg-split-bed/3",
"project": "admin/sbg-public-data",
"name": "SBG Split BED"
},
{
"href": "https://cgc-api.sbgenomics.com/v2/apps/admin/sbg-public-data/sbg-untar-fasta/8",
"id": "admin/sbg-public-data/sbg-untar-fasta/8",
"project": "admin/sbg-public-data",
"name": "SBG Untar fasta"
},
<snip>
],
"links": [
{
"href": "https://cgc-api.sbgenomics.com/v2/apps?visibility=public&offset=100&limit=100",
"rel": "next",
"method": "GET"
}
]
}
Scrolling through this list of apps, you'll see that RNA-seq Alignment - STAR isn't on this list of the first 100 results. To page through to the next 100 results, follow the path at the bottom of your response, "href": "https://cgc-api.sbgenomics.com/v2/apps?visibility=public&offset=100&limit=100". Use this path to issue another request which lists next 100 results starting from the 101st result, as shown below.
If RNA-seq Alignment - STAR is not in the results, page through until you see the workflow. Locate and copy the id of the RNA-seq Alignment - STAR workflow. We'll use this in the next step.
{
"href": "https://cgc-api.sbgenomics.com/v2/apps?visibility=public&offset=200&limit=100",
"items": [
<snip>
{
"href": "https://cgc-api.sbgenomics.com/v2/apps/admin/sbg-public-data/rna-seq-alignment-star/16",
"id": "admin/sbg-public-data/rna-seq-alignment-star/16",
"project": "admin/sbg-public-data",
"name": "RNA-seq Alignment - STAR"
},
<snip>
{
"href": "https://cgc-api.sbgenomics.com/v2/apps?visibility=public&offset=100&limit=100",
"rel": "prev",
"method": "GET"
}
]
}
We've located the id for the RNA-seq Alignment - STAR workflow, and now we can copy the workflow into our project.
###5b: Copy a workflow into a project
We can use the id we obtained above to copy the workflow into our project.
To copy a workflow, make the API request to copy an app, as shown below. Be sure to pass the app's id in the path of the request.
In the body of the request, include the name of the project you want to copy the public app into, such as rfranklin/batch-tutorial.
POST /v2/apps/admin/sbg-public-data/rna-seq-alignment-star/16/actions/copy HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
{
"project":"rfranklin/batch-tutorial"
}
This call returns the name and the id of the app within your project. Copy this id as we'll need it in the next step.
The response body also contains the full Common Workflow Language description of the copied app. This is typically a lengthy JSON object (raw), which we have omitted in part below for brevity. Keep this JSON handy for the next step, as it include information about setting up inputs for the workflow.
{
"href": "https://cgc-api.sbgenomics.com/v2/apps/rfranklin/batch-tutorial/RNA-seq Alignment - STAR/0",
"id": "rfranklin/batch-tutorial/rna-seq-alignment-star/0",
"project": "rfranklin/batch-tutorial",
"name": "RNA-seq Alignment - STAR",
"revision": 0,
<snip>
"inputs": [
{
"sbg:suggestedValue": [
{
"name": "Homo_sapiens.GRCh37.75.gtf",
"class": "File",
"path": "567890abc07c1752674486cd"
}
],
"id": "#sjdbGTFfile",
"label": "sjdbGTFfile",
"type": [
"null",
{
"items": "File",
"type": "array"
}
],
"sbg:y": 195.08331063389656,
"sbg:x": 160.49997586011762
},
{
"id": "#fastq",
"type": [
{
"items": "File",
"type": "array"
}
],
"label": "fastq",
"sbg:includeInPorts": true,
"sbg:y": 323.74995018542,
"sbg:x": 164.24999140203002
},
{
"sbg:suggestedValue": {
"name": "human_g1k_v37_decoy.phiX174_Homo_sapiens.GRCh37.75_star-2.4.2a.tar",
"class": "File",
"path": "567890abc07c17b56d99b0d6"
},
"id": "#genomeFastaFiles",
"label": "genomeFastaFiles",
"type": [
"File"
],
"sbg:y": 469.9999105781354,
"sbg:x": 167.749960079791
}
],
<snip>
}
We're now ready to modify the workflow we've copied.
###5c: Modify a workflow on the visual interface
After you copy a workflow from the Public Apps repository to your own project, you can edit the workflow via the visual interface or by editing its CWL. To edit the workflow, let's use the Workflow Editor on the visual interface.
We want to modify the RNA-seq Alignment - STAR workflow we copied above to take BAM files as inputs.
To modify the workflow, follow the directions below. Note that the following directions apply to the visual interface of the CGC.
- Navigate to the Apps tab of your project and click the pencil icon next to your copied workflow. You'll be taken to the Workflow Editor, as shown below.
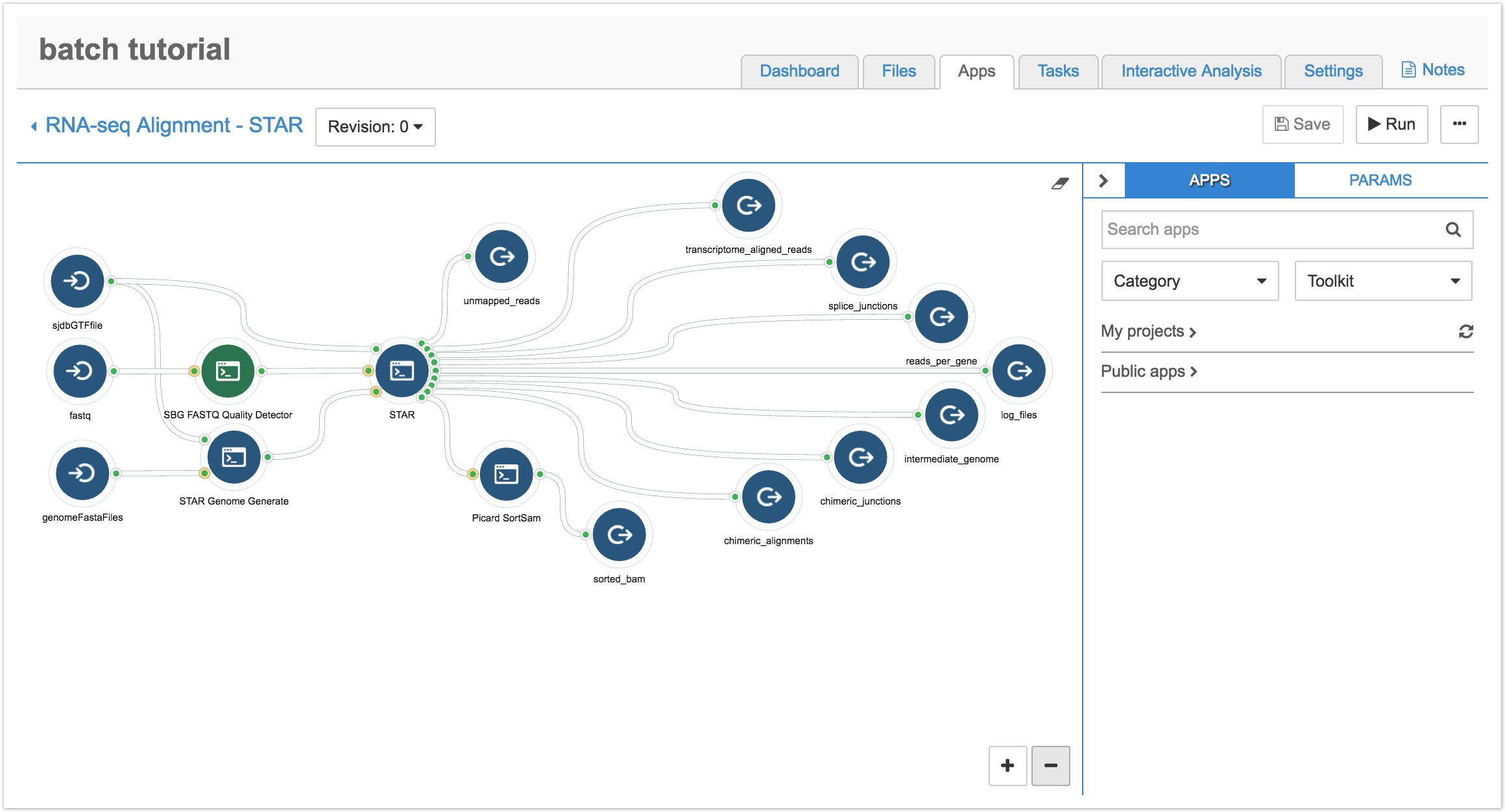
As shown above, the workflow has three input nodes (icons with arrows going into a half circle): sjdbGTFfile, fastq, and genomeFastaFiles. As our CCLE data is in the BAM format, we need to add the Picard SamToFastq tool so the workflow will accept BAM files by converting them to the fastq format.
- Click on the fastq input and select the red x above the node to delete it.
- Use the righthand APPS panel to search for the Picard SamToFastq tool.
- Drag and drop the Picard SamToFastq tool onto the Workflow Editor canvas.
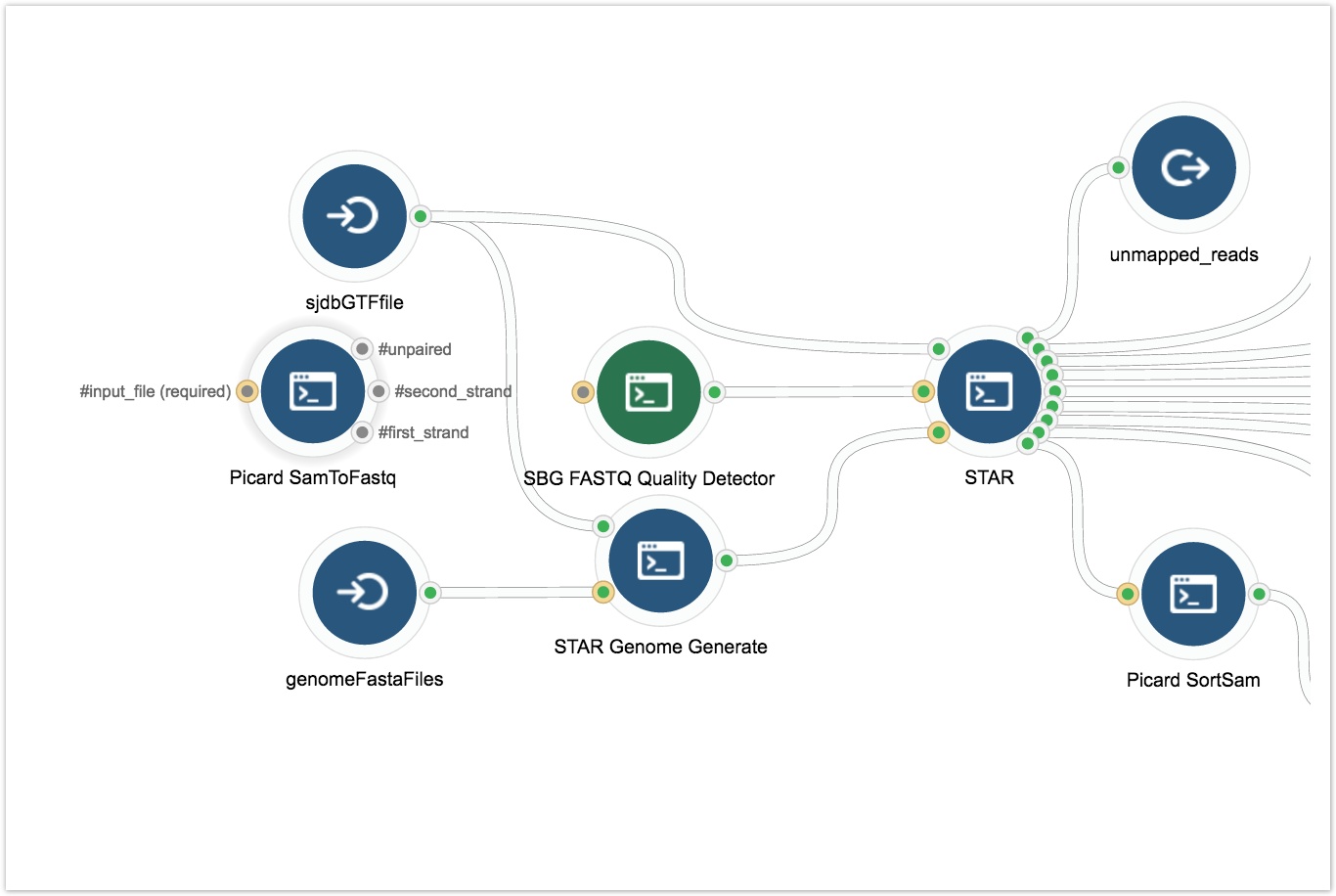
- Connect the Picard SAMToFastq tool to the SBG FASTQ Quality Detector tool as shown below.
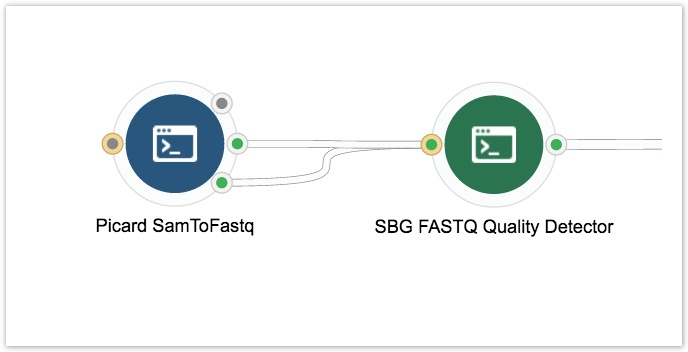
- Click on the circle on the left side of the Picard SamtoFastq tool and drag it out to the left side of the canvas. This adds an input node which accepts BAM files.
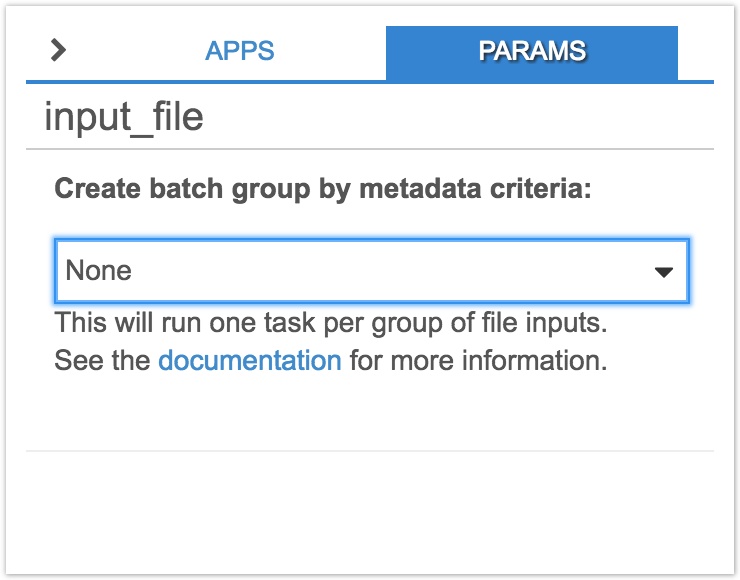
- Click on the input node, input_file. On the righthand panel, click on the dropdown menu below Create batch group by metadata criteria and select Sample ID. Note down the name of the input node, input_file. We'll use this information below to specify the input on which to batch when creating a draft task below
- Click Save in the top righthand corner and add an optional description, as shown below. Finish by clicking Save once more.
The modified RNA-seq Alignment - STAR workflow now accepts BAM inputs.
###5d: Find an app in your project via the API
Now that we've modified our workflow, we will return to the API to find the id for our modified workflow. To do this, we make the API request to list all apps available in a project. As shown in the example request below, supply a project id, such as rfranklin/batch-tutorial.
GET /v2/apps?project=rfranklin/batch-tutorial HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
This call returns the name and the id of the app within your project. Copy this id as we'll need it in the next step.
{
"href": "https://cgc-api.sbgenomics.com/v2/apps?offset=0&limit=1&project=rfranklin/batch-tutorial",
"items": [
{
"href": "https://cgc-api.sbgenomics.com/v2/apps/rfranklin/batch-tutorial/rna-seq-alignment-star/1",
"id": "rfranklin/batch-tutorial/rna-seq-alignment-star/1",
"project": "rfranklin/batch-tutorial",
"name": "RNA-seq Alignment - STAR"
}
],
"links": []
}
We're now ready to set up a draft batch task.
An app execution is called a task. Each task is associated with a set of input files and chosen settings for the tool(s) in the app. The first step to executing a task is to set up a draft task. In this step, you specify the inputs for your task.
To set up a draft task, make the API request to create a draft task, as shown below.
In the body of the request, specify the following:
name | A name for your task. |
app | The workflow you're running by including the workflow's id, which we obtained in the step above. |
batch_input | The input port on which you wish to batch, such as input_file from above. |
batch_by | The criteria on which to batch for the batch_by key. This consists of a type as well as the criteria. As shown below, we supply the first key, type, with the value, CRITERIA. We then supply the second key, criteria, with the value, metadata.sample_id. In short, we're batching by sample. |
project | Your project id, such as rfranklin/batch-tutorial. |
input | The reference files and data files for our workflow. Include the class, the file's path, and the file's name. The path is the file's id in our project. We obtained these ids when we copied the files to our project. |
An example request to create a draft task is as follows:
POST /v2/tasks HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
{
"name": "api batch tutorial task",
"app": "rfranklin/batch-tutorial/rna-seq-alignment-star/1",
"project": "rfranklin/batch-tutorial",
"batch_input": "input_file",
"batch_by": {
"type": "CRITERIA",
"criteria": [
"metadata.sample_id"
]
},
"inputs": {
"input_file": [
{
"class": "File",
"path": "567890abc4b002eed2cbe64f",
"name": "G28034.MDA-MB-361.1.bam"
},
{
"class": "File",
"path": "567890abc4b002eed2cbe64d",
"name": "G30603.TUHR4TKB.1.bam"
},
{
"class": "File",
"path": "567890abc4b002eed2cbe651",
"name": "G30630.VM-CUB1.3.bam"
}
],
"genomeFastaFiles": {
"class": "File",
"path": "567890abc4b002eed2cbf48b",
"name": "HG19_Broad_variant.fasta"
},
"sjdbGTFfile": [
{
"name": "Homo_sapiens.GRCh37.75.gtf",
"class": "File",
"path": "567890abc4b002eed2cbf489"
}
]
}
}
The response body will indicate if your draft task was successfully created. You'll also see the id for your draft task. Copy this to your clipboard, as we'll use it in the next step.
Note that you'll also see error messages if you've made a mistake in entering your inputs.
{
"href": "https://cgc-api.sbgenomics.com/v2/tasks/48f79ccf-12b3-45b6-789c-b1e8d88dabcd",
"id": "48f79ccf-12b3-45b6-789c-b1e8d88dabcd",
"name": "api batch tutorial task",
"status": "DRAFT",
"project": "rfranklin/batch-tutorial",
"app": "rfranklin/batch-tutorial/rna-seq-alignment-star/1",
"type": "v2",
"created_by": "rfranklin",
"start_time": "2016-09-23T18:43:24Z",
"batch": true,
"batch_input": "input_file",
"batch_by": {
"type": "CRITERIA",
"criteria": [
"metadata.sample_id"
]
},
"errors": [],
"warnings": [],
"inputs": {
<snip>
}
}
Note that batch is set to true. This indicates we've created a draft batch task. The response also reiterates the criteria by which we're batching. Now, we're ready to run the task.
To run a task on the CGC, you'll need your draft task's id, obtained in the step above. Then, make the API request to run a task, specifying the id of the draft task, as shown below.
POST /v2/tasks/48f79ccf-12b3-45b6-789c-b1e8d88dabcd/actions/run" HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
Your response body will contain information about your task as well as its status. Learn more about what happens when you run a task.
{
"href": "https://cgc-api.sbgenomics.com/v2/tasks/48f79ccf-12b3-45b6-789c-b1e8d88dabcd",
"id": "48f79ccf-12b3-45b6-789c-b1e8d88dabcd",
"name": "api batch tutorial task",
"status": "CREATING",
"project": "rfranklin/batch-tutorial",
"app": "rfranklin/batch-tutorial/rna-seq-alignment-star/1",
"type": "v2",
"created_by": "rfranklin",
"executed_by": "rfranklin",
"start_time": "2016-09-23T18:43:24Z",
"batch": true,
"batch_input": "input_file",
"batch_by": {
"type": "CRITERIA",
"criteria": [
"metadata.sample_id"
]
},
"errors": [],
"warnings": [],
"inputs": {
<snip>
},
}
Copy down the id of the parent task from the response body. You'll use this in the next step to obtain your task outputs. You'll be notified by email once your task has completed.
Once your task has completed, you can get your task outputs. First, we obtain the task ids of the child tasks. Then, we make the API request to get details of a task.
Before we can obtain the outputs of each child task, we have to obtain their task ids by making the API request to list tasks. To list all tasks associated with a parent task, set the parameter parent to the task id of the parent task obtained in the step above. For instance, in the example below, parent is set to e3a88ab3-829a-485c-995a-3acd817ee98c.
GET /v2/tasks?parent=48f79ccf-12b3-45b6-789c-b1e8d88dabcd HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
The response body returns a list of the child tasks, including the id of each child task, the name of the parent task, and the project the tasks belong to. Copy the id of each child task to a clipboard. We'll use this in the next step.
{
"href": "https://cgc-api.sbgenomics.com/v2/tasks?parent=48f79ccf-12b3-45b6-789c-b1e8d88dabcd&offset=0&limit=3",
"items": [
{
"href": "https://cgc-api.sbgenomics.com/v2/tasks/1fd125fa-789c-45b6-12b3-2a3ab3bedcba",
"id": "1fd125fa-789c-45b6-12b3-2a3ab3bedcba",
"name": "api batch tutorial task",
"project": "rfranklin/batch-tutorial"
},
{
"href": "https://cgc-api.sbgenomics.com/v2/tasks/f0b89de2-45b6-789c-12b3-05b832c576c6",
"id": "f0b89de2-45b6-789c-12b3-05b832c576c6",
"name": "api batch tutorial task",
"project": "rfranklin/batch-tutorial"
},
{
"href": "https://cgc-api.sbgenomics.com/v2/tasks/67f68072-45b6-12b3-789c-37be8b0f2f04",
"id": "67f68072-45b6-12b3-789c-37be8b0f2f04",
"name": "api batch tutorial task",
"project": "rfranklin/batch-tutorial"
}
],
"links": []
}
###8b: Obtain the outputs of a child task
For each child task above, make the following API request to get details of a task to obtain its outputs. Be sure to pass along the child task's id, obtained in the step above, in the path. Add the parameter fields=outputs to filter the response body to only display the outputs, as shown below.
GET /v2/tasks/67f68072-45b6-12b3-789c-37be8b0f2f04?fields=outputs HTTP/1.1
Host: cgc-api.sbgenomics.com
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
The response body returns the outputs of your task, including file ids (path) in case you wish to perform further analyses on these files.
{
"outputs": {
"log_files": [
{
"path": "567890abc4b0ebec9056ec72",
"name": "G30603.TUHR4TKB.1.converted.pe.Log.final.out",
"class": "File"
},
{
"path": "567890abc4b0b3cd0ec80c15",
"name": "G30603.TUHR4TKB.1.converted.pe.Log.out",
"class": "File"
},
{
"path": "567890abc4b0ebec9056ec74",
"name": "G30603.TUHR4TKB.1.converted.pe.Log.progress.out",
"class": "File"
}
],
"reads_per_gene": {},
"unmapped_reads": [
{
"path": "567890abc4b0b3cd0ec80c17",
"name": "G30603.TUHR4TKB.1.converted.pe.Unmapped.out.mate1.fastq",
"class": "File"
},
{
"path": "567890abc4b0ebec9056ec76",
"name": "G30603.TUHR4TKB.1.converted.pe.Unmapped.out.mate2.fastq",
"class": "File"
}
],
"chimeric_junctions": {},
"sorted_bam": {
"path": "567890abc4b0ebec9056f2c0",
"name": "G30603.TUHR4TKB.1.converted.pe.Aligned.out.sorted.bam",
"secondaryFiles": [
{
"path": "567890abc4b0b3cd0ec810ef",
"size": 4018336,
"name": "G30603.TUHR4TKB.1.converted.pe.Aligned.out.sorted.bam.bai",
"class": "File"
}
],
"class": "File"
},
"chimeric_alignments": {},
"transcriptome_aligned_reads": {
"path": "567890abc4b0b3cd0ec80c19",
"name": "G30603.TUHR4TKB.1.converted.pe.Aligned.toTranscriptome.out.bam",
"class": "File"
},
"intermediate_genome": {},
"splice_junctions": {
"path": "567890abc4b0ebec9056ec78",
"name": "G30603.TUHR4TKB.1.converted.pe.SJ.out.tab",
"class": "File"
}
}
}
Repeat this process for the other child tasks to obtain their respective outputs.
That’s it! We've executed a batch analysis and obtained some results.
Updated less than a minute ago
