QUERY DATASETS
Overview
Build queries to locate a subset of data within a dataset for further analysis. Queries use metadata properties and their values to filter datasets for specific entities. The CGC provides the following two query methods:
- Method one: the Data Browser - Use the Data Browser to query datasets via an intuitive visual interface. The Data Browser's autosuggest and search functionalities facilitate querying while alleviating the need to learn new terminology.
- Method two: the Datasets API - Use the Datasets API to programmatically browse and query datasets using API requests written in JSON. The Datasets API is suitable for queries containing numerous parameters since the query is formatted as a concise dictionary.
Comparing the three query methods
In this section, we query the TCGA dataset for file entities and use metadata properties to filter results to file entities which come from RNA-Seq analyses of cases that have the vital status "alive," a gender of "female," and a diagnosis of "breast cancer". This query is reproduced three times below, once in each of the three query methods.
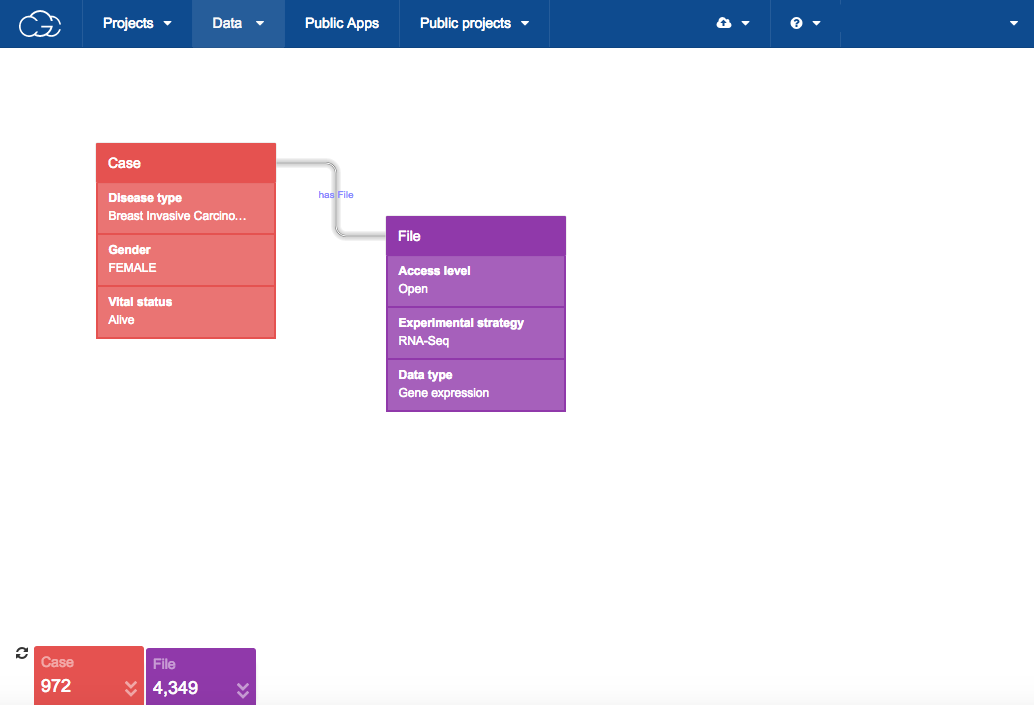
Method one: the Data Browser
Use the Data Browser to construct a query in the CGC visual interface.
The query shown below starts from the Case entity and provides specific values for its metadata properties of Disease type, Gender, and Vital status. For example, Gender has a value of FEMALE. The query below also contains a File entity with specific values for Access level, Experimental strategy, and Data type. The File entity designates that we are looking for files for the cases that match the specified criteria.
Below the query, refreshable count cards reveal that 972 cases and 4,349 files match the query criteria.
Learn more about the Data Browser.
Method two: the Datasets API
Query datasets programmatically via a JSON request using the Datasets API.
The query below searches for members of the files entity with the following metadata properties: an access level of Open, a data type of Gene expression, and an experimental strategy of RNA-Seq. These files are for cases with a disease type of Breast Invasive Carcinoma, a gender of FEMALE, and a vital status of Alive.
Learn more about the Datasets API.
POST datasets/tcga/v0/query HTTP/1.1
Host: cgc-datasets-api.sbgenomics.com
X-SBG-Auth-Token: 3210a98c1db9304ea9d9273156740f74
{
"entity": "files",
"hasAccessLevel" : "Open",
"hasDataType" : "Gene expression",
"hasExperimentalStrategy": "RNA-Seq",
"hasCase": {
"hasDiseaseType" : "Breast Invasive Carcinoma",
"hasGender" : "FEMALE",
"hasVitalStatus" : "Alive"
}
}
Resources
Updated 7 months ago
