↳ Case Explorer features
BROWSE DATASETS > About the Case Explorer > Case Explorer features
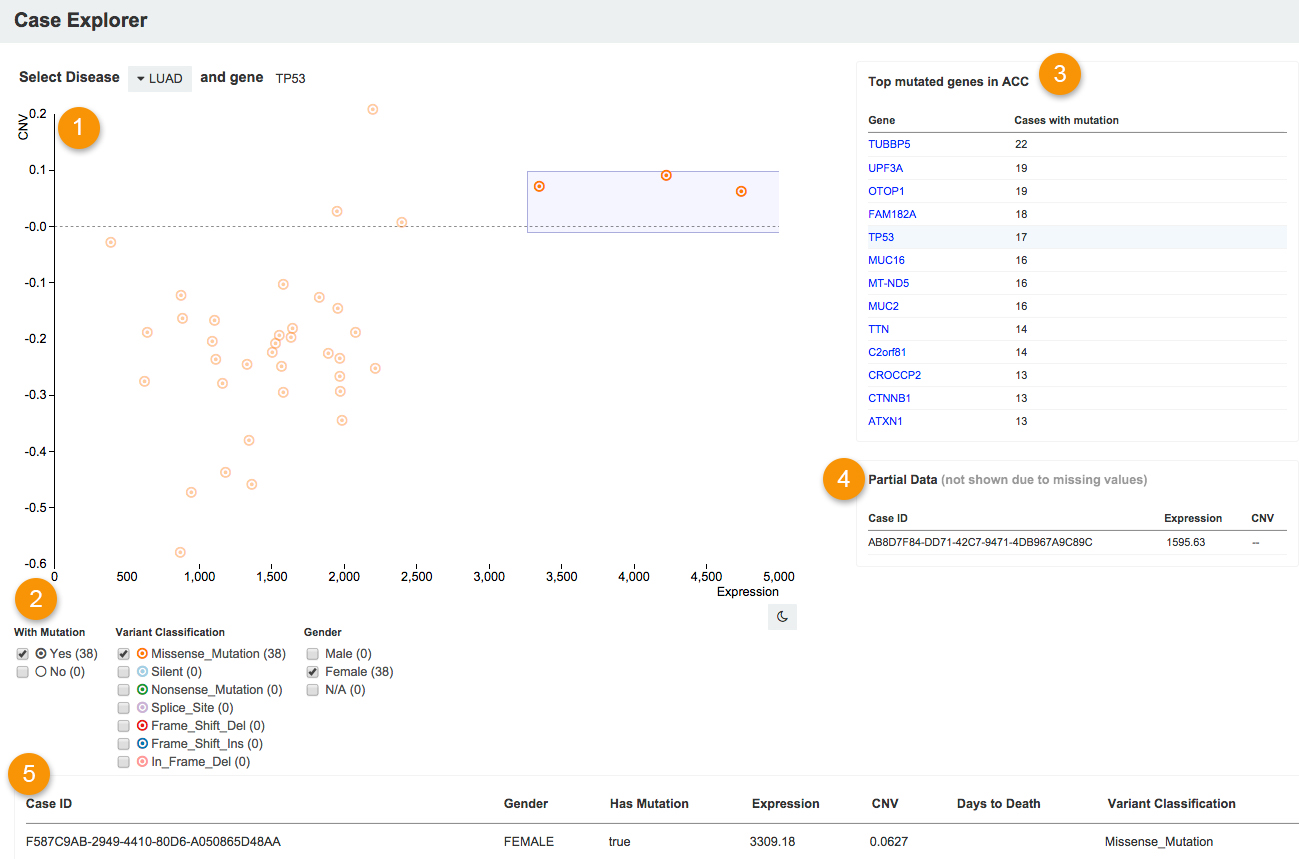
##[ 1 ] Scatter plot
The scatter plot in the Case Explorer shows the relation between copy number variation (CNV) on the y-axis and gene expression levels on the x-axis for a chosen gene in patients with a specific type of cancer.
CNV is calculated using the Circular binary segmentation (CBS) algorithm. The data shows the mean log2 ratio of the number of probes on the array that are represented in the region containing the chosen gene, centered on 0. The probes are sorted based on the order of hg19 reference genome. All values are taken from the level 3 *.hg19.seg.txt files for tumor samples.
Gene expression is calculated using the RSEM software package. The data represents gene abundance estimates normalized to set the upper quartile count at 1000. All values are taken from the level 3 unc.edu.[sample id].rsem.genes.normalized_results files for tumor samples.
Top Mutated Genes per disease
You can see the Top Mutated Genes for each disease in a separate table. Click on one of the genes to see them in the Case Explorer.
Each circle on the scatter plot represents a single case (patient). Empty gray circles represent cases where the gene is not mutated. Circles with dots show cases with mutated genes. In addition, each type of mutation is differentiated by color. For instance, silent mutations are light blue, missense mutations are orange, and frame shift deletions are red. The color code for the mutations from the Variant Classification table is below the scatter plot.
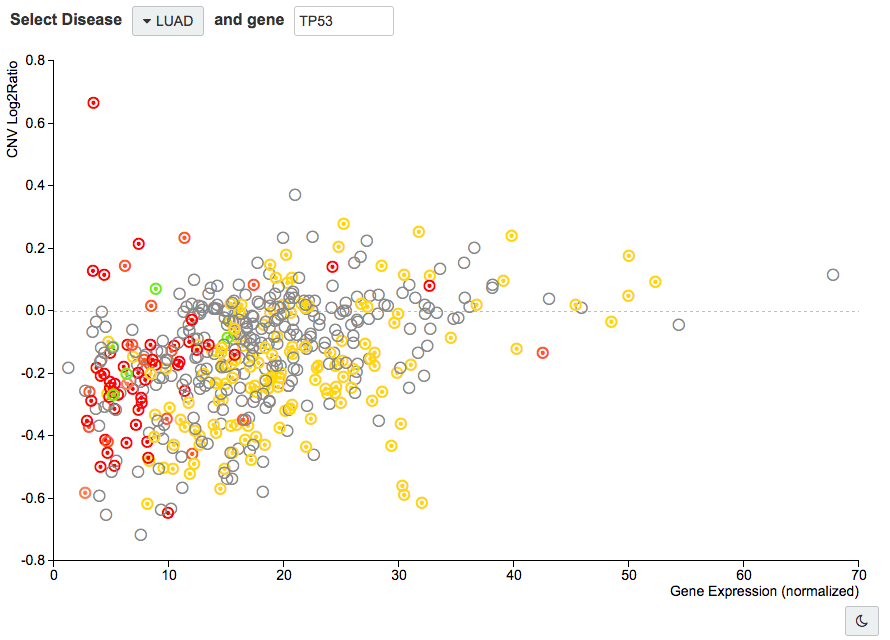
Night mode
To help you visualize the cases, you can also toggle the default scatter plot look to night mode. Click the moon icon in the bottom right corner.
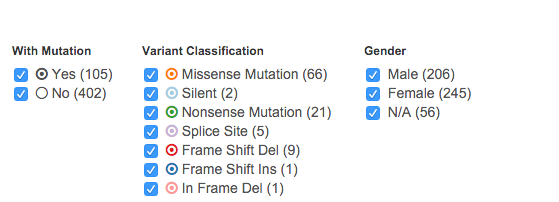
[ 2 ] Filters
Below the scatter plot, three filters help you further refine your data by Mutation status, Variant Classification (Mutation type), and Gender. Numbers next to filter names represent number of cases with the specific characteristic. Not all mutation types are constantly displayed. The Variant Classification and Gender lists dynamically update based on the selected disease and gene.
[ 3 ] Top Mutated Genes table
The most highly mutated genes for the selected disease are displayed in the Top Mutated Genes table.
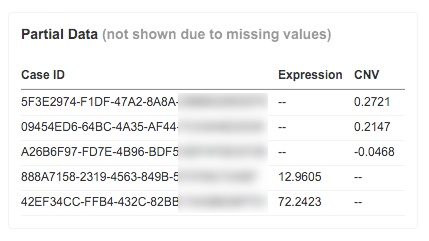
[ 4 ] Partial Data table
This table shows all the cases which are not represented in the Case Explorer due to missing data, such as gene expression values or CNV data (or both). The Partial Data table lists the values these cases are missing.
[ 5 ] Selected Cases table
Once cases are selected on the scatter plot, they will be displayed in a table below the scatter plot. Case IDs as well as exact expression levels and CNV values are also shown.
To export these cases, along with all the related information, to a CSV file, click Export as CSV below the table. To further analyze this data, click Continue to Data Browser.
Updated over 1 year ago
